Lipid synthesis and transport are coupled to regulate membrane lipid dynamics in the endoplasmic reticulum
- PMID: 31108203
- PMCID: PMC6858525
- DOI: 10.1016/j.bbalip.2019.05.005
Lipid synthesis and transport are coupled to regulate membrane lipid dynamics in the endoplasmic reticulum
Abstract
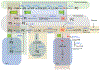
Structural lipids are mostly synthesized in the endoplasmic reticulum (ER), from which they are actively transported to the membranes of other organelles. Lipids can leave the ER through vesicular trafficking or non-vesicular lipid transfer and, curiously, both processes can be regulated either by the transported lipid cargos themselves or by different secondary lipid species. For most structural lipids, transport out of the ER membrane is a key regulatory component controlling their synthesis. Distribution of the lipids between the two leaflets of the ER bilayer or between the ER and other membranes is also critical for maintaining the unique membrane properties of each cellular organelle. How cells integrate these processes within the ER depends on fine spatial segregation of the molecular components and intricate metabolic channeling, both of which we are only beginning to understand. This review will summarize some of these complex processes and attempt to identify the organizing principles that start to emerge. This article is part of a Special Issue entitled Endoplasmic reticulum platforms for lipid dynamics edited by Shamshad Cockcroft and Christopher Stefan.
Keywords: Endoplasmic reticulum; Lipid transfer protein; Membrane contact sites; Non-vesicular lipid transfer; Phosphatidylcholine; Phosphatidylinositol; Phosphatidylserine.
Published by Elsevier B.V.
Conflict of interest statement
The Authors declare no conflict of interest related to this work.
Figures

References
-
- Kent C, Eukaryotic phospholipid biosynthesis, Annu Rev Biochem, 64 (1995) 315–343. - PubMed
-
- Gonzalez-Baro MR, Coleman RA, Mitochondrial acyltransferases and glycerophospholipid metabolism, Biochim Biophys Acta, 1862 (2017) 49–55. - PubMed
-
- Wilfling F, Wang H, Haas JT, Krahmer N, Gould TJ, Uchida A, Cheng JX, Graham M, Christiano R, Frohlich F, Liu X, Buhman KK, Coleman RA, Bewersdorf J, Farese RV Jr., Walther TC, Triacylglycerol synthesis enzymes mediate lipid droplet growth by relocalizing from the ER to lipid droplets, Dev Cell, 24 (2013) 384–399. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

