IL-1 beta-mediated macrophage-hepatocyte crosstalk upregulates hepcidin under physiological low oxygen levels
- PMID: 31108461
- PMCID: PMC6526398
- DOI: 10.1016/j.redox.2019.101209
IL-1 beta-mediated macrophage-hepatocyte crosstalk upregulates hepcidin under physiological low oxygen levels
Abstract
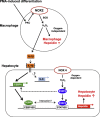
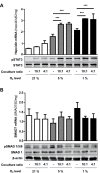
In mammals, the iron masterswitch hepcidin efficiently controls iron recycling by the macrophage-liver axis but the exact interplay between macrophages and hepatocytes remains poorly understood. We here study hepcidin response during macrophage differentiation as well as the macrophage-hepatocyte crosstalk and its subsequent effects on hepatocyte hepcidin using an in vitro co-culture model that mimics the physiological liver microenvironment. We show that macrophage differentiation strongly induces hepcidin by 60-fold both in THP1 macrophages and primary isolated monocyte-derived macrophages. Removal of H2O2 by catalase or inhibition of NOX2 efficiently blocked hepcidin induction. After differentiation, macrophage hepcidin accounted for 10% of total hepatocyte hepcidin and did not respond to low oxygen levels. In contrast, co-culture of differentiated macrophages with Huh7 cells significantly induced hepatocyte hepcidin, which was further potentiated under low oxygen levels. Hepatocyte hepcidin was also upregulated when Huh7 cells were solely exposed to macrophage-conditioned hypoxic medium. A cytokine screen identified macrophage secreted IL-1β as major inducer of hepcidin in hepatocytes. In confirmation, treatment of Huh7 cells with the IL-1 receptor antagonist (anakinra) completely blunted macrophage-mediated hepcidin transcription in hepatocytes. Finally, detailed analysis of potentially involved signaling pathways points toward STAT3 and CEBPδ-mediated hepcidin induction independent of IL-6. In conclusion, our study demonstrates a strong NOX2-mediated hepcidin induction during macrophage differentiation. These differentiated macrophages are able to efficiently induce hepatocyte hepcidin mainly through secretion of IL-1β. Our data highlight a hitherto unrecognized role of macrophage-hepatocyte crosstalk for a joint and oxygen-dependent hepcidin production through STAT3 and CEBPδ.
Keywords: Cytokines; Hydrogen peroxide; Hypoxia; Iron metabolism/hepcidin; NADPH oxidase; STAT3.
Copyright © 2019 The Authors. Published by Elsevier B.V. All rights reserved.
Figures






Similar articles
-
Hypoxia enhances H2O2-mediated upregulation of hepcidin: Evidence for NOX4-mediated iron regulation.Redox Biol. 2018 Jun;16:1-10. doi: 10.1016/j.redox.2018.02.005. Epub 2018 Feb 12. Redox Biol. 2018. PMID: 29459227 Free PMC article.
-
Interleukin-1β (IL-1β) transcriptionally activates hepcidin by inducing CCAAT enhancer-binding protein δ (C/EBPδ) expression in hepatocytes.J Biol Chem. 2017 Jun 16;292(24):10275-10287. doi: 10.1074/jbc.M116.770974. Epub 2017 Apr 24. J Biol Chem. 2017. PMID: 28438835 Free PMC article.
-
Activated macrophages induce hepcidin expression in HuH7 hepatoma cells.Haematologica. 2009 Jun;94(6):773-80. doi: 10.3324/haematol.2008.003400. Epub 2009 May 19. Haematologica. 2009. PMID: 19454498 Free PMC article.
-
Hepcidin and IL-1β.Vitam Horm. 2019;110:143-156. doi: 10.1016/bs.vh.2019.01.007. Epub 2019 Feb 2. Vitam Horm. 2019. PMID: 30798809 Review.
-
Beyond anemia: hepcidin, monocytes and inflammation.Biol Chem. 2013 Feb;394(2):231-8. doi: 10.1515/hsz-2012-0217. Biol Chem. 2013. PMID: 23314535 Review.
Cited by
-
dECM restores macrophage immune homeostasis and alleviates iron overload to promote DTPI healing.Regen Biomater. 2024 Jan 17;11:rbad118. doi: 10.1093/rb/rbad118. eCollection 2024. Regen Biomater. 2024. PMID: 38404617 Free PMC article.
-
Basic biology and roles of CEBPD in cardiovascular disease.Cell Death Discov. 2025 Mar 14;11(1):102. doi: 10.1038/s41420-025-02357-4. Cell Death Discov. 2025. PMID: 40087290 Free PMC article. Review.
-
CCAAT/Enhancer-Binding Proteins in Fibrosis: Complex Roles Beyond Conventional Understanding.Research (Wash D C). 2022 Oct 3;2022:9891689. doi: 10.34133/2022/9891689. eCollection 2022. Research (Wash D C). 2022. PMID: 36299447 Free PMC article. Review.
-
Impaired inflammasome activation and bacterial clearance in G6PD deficiency due to defective NOX/p38 MAPK/AP-1 redox signaling.Redox Biol. 2020 Jan;28:101363. doi: 10.1016/j.redox.2019.101363. Epub 2019 Nov 2. Redox Biol. 2020. PMID: 31707353 Free PMC article.
-
Inflammatory adipose activates a nutritional immunity pathway leading to retinal dysfunction.Cell Rep. 2022 Jun 14;39(11):110942. doi: 10.1016/j.celrep.2022.110942. Cell Rep. 2022. PMID: 35705048 Free PMC article.
References
-
- Krause A., Neitz S., Magert H.J., Schulz A., Forssmann W.G., Schulz-Knappe P., Adermann K. LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS Lett. 2000;480(2–3):147–150. - PubMed
- A. Krause, S. Neitz, H.J. Magert, A. Schulz, W.G. Forssmann, P. Schulz-Knappe, K. Adermann, LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity, FEBS Lett 480(2-3) (2000) 147-150. - PubMed
-
- Park C.H., Valore E.V., Waring A.J., Ganz T. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J. Biol. Chem. 2001;276(11):7806–7810. - PubMed
- C.H. Park, E.V. Valore, A.J. Waring, T. Ganz, Hepcidin, a urinary antimicrobial peptide synthesized in the liver, J Biol Chem 276(11) (2001) 7806-7810. - PubMed
-
- Sow F.B., Florence W.C., Satoskar A.R., Schlesinger L.S., Zwilling B.S., Lafuse W.P. Expression and localization of hepcidin in macrophages: a role in host defense against tuberculosis. J. Leukoc. Biol. 2007;82(4):934–945. - PubMed
- F.B. Sow, W.C. Florence, A.R. Satoskar, L.S. Schlesinger, B.S. Zwilling, W.P. Lafuse, Expression and localization of hepcidin in macrophages: a role in host defense against tuberculosis, J Leukoc Biol 82(4) (2007) 934-945. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

