Genetic Variability and Evolution of Hepatitis E Virus
- PMID: 31109076
- PMCID: PMC6563261
- DOI: 10.3390/v11050456
Genetic Variability and Evolution of Hepatitis E Virus
Abstract
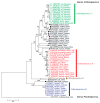
Hepatitis E virus (HEV) is a single-stranded positive-sense RNA virus. HEV can cause both acute and chronic hepatitis, with the latter usually occurring in immunocompromised patients. Modes of transmission range from the classic fecal-oral route or zoonotic route, to relatively recently recognized but increasingly common routes, such as via the transfusion of blood products or organ transplantation. Extrahepatic manifestations, such as neurological, kidney and hematological abnormalities, have been documented in some limited cases, typically in patients with immune suppression. HEV has demonstrated extensive genomic diversity and a variety of HEV strains have been identified worldwide from human populations as well as growing numbers of animal species. The genetic variability and constant evolution of HEV contribute to its physiopathogenesis and adaptation to new hosts. This review describes the recent classification of the Hepeviridae family, global genotype distribution, clinical significance of HEV genotype and genomic variability and evolution of HEV.
Keywords: evolution; genetic variability; genotypes; hepatitis E virus.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

