Profiling of Small Molecular Metabolites in Nostoc flagelliforme during Periodic Desiccation
- PMID: 31109094
- PMCID: PMC6562405
- DOI: 10.3390/md17050298
Profiling of Small Molecular Metabolites in Nostoc flagelliforme during Periodic Desiccation
Abstract
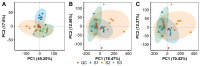
The mass spectrometry-based metabolomics approach has become a powerful tool for the quantitative analysis of small-molecule metabolites in biological samples. Nostoc flagelliforme, an edible cyanobacterium with herbal value, serves as an unexploited bioresource for small molecules. In natural environments, N. flagelliforme undergoes repeated cycles of rehydration and dehydration, which are interrupted by either long- or short-term dormancy. In this study, we performed an untargeted metabolite profiling of N. flagelliforme samples at three physiological states: Dormant (S1), physiologically fully recovered after rehydration (S2), and physiologically partially inhibited following dehydration (S3). Significant metabolome differences were identified based on the OPLS-DA (orthogonal projections to latent structures discriminant analysis) model. In total, 183 differential metabolites (95 up-regulated; 88 down-regulated) were found during the rehydration process (S2 vs. S1), and 130 (seven up-regulated; 123 down-regulated) during the dehydration process (S3 vs. S2). Thus, it seemed that the metabolites' biosynthesis mainly took place in the rehydration process while the degradation or possible conversion occurred in the dehydration process. In addition, lipid profile differences were particularly prominent, implying profound membrane phase changes during the rehydration-dehydration cycle. In general, this study expands our understanding of the metabolite dynamics in N. flagelliforme and provides biotechnological clues for achieving the efficient production of those metabolites with medical potential.
Keywords: LC-MS; Nostoc flagelliforme; cyanobacteria; metabolic profiling; metabolites; rehydration and dehydration.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Lankadurai B.P., Nagato E.G., Simpson M.J. Environmental metabolomics: An emerging approach to study organism responses to environmental stressors. Environ. Rev. 2013;21:180–205. doi: 10.1139/er-2013-0011. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

