How fall dormancy benefits alfalfa winter-survival? Physiologic and transcriptomic analyses of dormancy process
- PMID: 31109303
- PMCID: PMC6528297
- DOI: 10.1186/s12870-019-1773-3
How fall dormancy benefits alfalfa winter-survival? Physiologic and transcriptomic analyses of dormancy process
Abstract
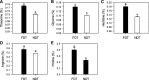
Background: Fall dormancy and freezing tolerance characterized as two important phenotypic traits, have great effects on productivity and persistence of alfalfa (Medicago sativa L.). Despite the fact that one of the most limiting traits for alfalfa freezing tolerance in winter is fall dormancy, the interplay between fall dormancy and cold acclimation processes of alfalfa remains largely unknown. We compared the plant regrowth, winter survival, raffinose and amino acids accumulation, and genome-wide differentially expressed genes of fall-dormant cultivar with non-dormant cultivar under cold acclimation.
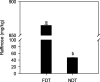
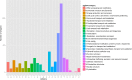
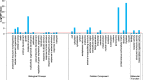
Results: Averaged over both years, the non-dormant alfalfa exhibited largely rapid regrowth compared with fall dormant alfalfa after last cutting in autumn, but the winter survival rate of fall dormant alfalfa was about 34-fold higher than that of non-dormant alfalfa. The accumulation of raffinose and amino acids were significantly increased in fall dormant alfalfa, whereas were decreased in non-dormant alfalfa under cold acclimation. Expressions of candidate genes encoding raffinose biosynthesis genes were highly up-regulated in fall dormant alfalfa, but down-regulated in non-dormant alfalfa under cold acclimation. In fall dormant alfalfa, there was a significantly down-regulated expression of candidate genes encoding the glutamine synthase, which is indirectly involved in the proline metabolism. A total of eight significantly differentially expressed transcription factors (TFs) related to CBF and ABRE-BFs were identified. The most up-regulated TFs in fall dormant alfalfa cultivar were ABF4 and DREB1C.
Conclusions: Fall dormant alfalfa drastically increased raffinose and amino acids accumulation under cold acclimation. Raffinose-associated and amino acid-associated genes involved in metabolic pathways were more highly expressed in fall dormant alfalfa than non-dormant alfalfa under cold acclimation. This global survey of transcriptome profiles provides new insights into the interplay between fall dormancy and cold acclimation in alfalfa.
Keywords: Fall dormancy; Medicago sativa; RNA-Seq; Raffinose and amino acids; qRT-PCR.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
De novo characterization of fall dormant and nondormant alfalfa (Medicago sativa L.) leaf transcriptome and identification of candidate genes related to fall dormancy.PLoS One. 2015 Mar 23;10(3):e0122170. doi: 10.1371/journal.pone.0122170. eCollection 2015. PLoS One. 2015. PMID: 25799491 Free PMC article.
-
Effect of photoperiod prior to cold acclimation on freezing tolerance and carbohydrate metabolism in alfalfa (Medicago sativa L.).Plant Sci. 2017 Nov;264:122-128. doi: 10.1016/j.plantsci.2017.09.003. Epub 2017 Sep 12. Plant Sci. 2017. PMID: 28969792
-
Genome-wide identification of different dormant Medicago sativa L. MicroRNAs in response to fall dormancy.PLoS One. 2014 Dec 4;9(12):e114612. doi: 10.1371/journal.pone.0114612. eCollection 2014. PLoS One. 2014. PMID: 25473944 Free PMC article.
-
Deacclimation after cold acclimation-a crucial, but widely neglected part of plant winter survival.J Exp Bot. 2019 Sep 24;70(18):4595-4604. doi: 10.1093/jxb/erz229. J Exp Bot. 2019. PMID: 31087096 Free PMC article. Review.
-
The potential for an increasing threat of unseasonal temperature cycles to dormant plants.New Phytol. 2024 Oct;244(2):377-383. doi: 10.1111/nph.20052. Epub 2024 Aug 17. New Phytol. 2024. PMID: 39152704 Review.
Cited by
-
A Pan-Transcriptome Analysis Indicates Efficient Downregulation of the FIB Genes Plays a Critical Role in the Response of Alfalfa to Cold Stress.Plants (Basel). 2022 Nov 17;11(22):3148. doi: 10.3390/plants11223148. Plants (Basel). 2022. PMID: 36432878 Free PMC article.
-
Genome-Wide Association Analysis Coupled With Transcriptome Analysis Reveals Candidate Genes Related to Salt Stress in Alfalfa (Medicago sativa L.).Front Plant Sci. 2022 Feb 3;12:826584. doi: 10.3389/fpls.2021.826584. eCollection 2021. Front Plant Sci. 2022. PMID: 35185967 Free PMC article.
-
The effect of alfalfa cultivation on improving physicochemical properties soil microorganisms community structure of grey desert soil.Sci Rep. 2023 Aug 23;13(1):13747. doi: 10.1038/s41598-023-41005-8. Sci Rep. 2023. PMID: 37612457 Free PMC article.
-
Physiological characteristics and transcriptomic analyses of alfalfa root crown in wintering.Front Plant Sci. 2024 Dec 9;15:1486564. doi: 10.3389/fpls.2024.1486564. eCollection 2024. Front Plant Sci. 2024. PMID: 39717732 Free PMC article.
-
Unveiling the Cold Acclimation of Alfalfa: Insights into Its Starch-Soluble Sugar Dynamic Transformation.Plants (Basel). 2025 Apr 26;14(9):1313. doi: 10.3390/plants14091313. Plants (Basel). 2025. PMID: 40364342 Free PMC article.
References
-
- Ariss JJ, Vandemark GJ. Assessment of genetic diversity among nondormant and semidormant alfalfa populations using sequence-related amplified polymorphisms. Crop Sci. 2007;47(6):2274. doi: 10.2135/cropsci2006.12.0782. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

