Genetic and epigenetic regulation of human aging and longevity
- PMID: 31109447
- PMCID: PMC7295568
- DOI: 10.1016/j.bbadis.2018.08.039
Genetic and epigenetic regulation of human aging and longevity
Abstract
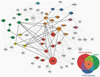
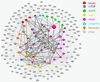
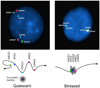
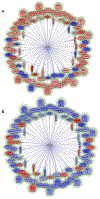
Here we summarize the latest data on genetic and epigenetic contributions to human aging and longevity. Whereas environmental and lifestyle factors are important at younger ages, the contribution of genetics appears more important in reaching extreme old age. Genome-wide studies have implicated ~57 gene loci in lifespan. Epigenomic changes during aging profoundly affect cellular function and stress resistance. Dysregulation of transcriptional and chromatin networks is likely a crucial component of aging. Large-scale bioinformatic analyses have revealed involvement of numerous interaction networks. As the young well-differentiated cell replicates into eventual senescence there is drift in the highly regulated chromatin marks towards an entropic middle-ground between repressed and active, such that genes that were previously inactive "leak". There is a breakdown in chromatin connectivity such that topologically associated domains and their insulators weaken, and well-defined blocks of constitutive heterochromatin give way to generalized, senescence-associated heterochromatin, foci. Together, these phenomena contribute to aging.
Keywords: APOE; Aging; Epigenetics; FOXO3; Genome-wide association studies; Histones; Longevity; Network analysis; Transcription.
Copyright © 2018. Published by Elsevier B.V.
Figures




References
-
- McGue M, Vaupel JW, Holm N, Harvald B, Longevity is moderately heritable in a sample of Danish twins born 1870–1880, J. Gerontol 48 (1993) B237–B244. - PubMed
-
- Philippe P, Familial correlations of longevity: an isolate-based study, Am. J. Med. Genet 2 (1978) 121–129. - PubMed
-
- Mayer PJ, Inheritance of longevity evinces no secular trend among members of six New England families born 1650–1874, Am. J. Hum. Biol 3 (1991) 49–58. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

