NPR1 Promotes Its Own and Target Gene Expression in Plant Defense by Recruiting CDK8
- PMID: 31110139
- PMCID: PMC6716257
- DOI: 10.1104/pp.19.00124
NPR1 Promotes Its Own and Target Gene Expression in Plant Defense by Recruiting CDK8
Abstract
NPR1 (NONEXPRESSER OF PR GENES1) functions as a master regulator of the plant hormone salicylic acid (SA) signaling and plays an essential role in plant immunity. In the nucleus, NPR1 interacts with transcription factors to induce the expression of PR (PATHOGENESIS-RELATED) genes and thereby promote defense responses. However, the underlying molecular mechanism of PR gene activation is poorly understood. Furthermore, despite the importance of NPR1 in plant immunity, the regulation of NPR1 expression has not been extensively studied. Here, we show that SA promotes the interaction of NPR1 with both CDK8 (CYCLIN-DEPENDENT KINASE8) and WRKY18 (WRKY DNA-BINDING PROTEIN18) in Arabidopsis (Arabidopsis thaliana). NPR1 recruits CDK8 and WRKY18 to the NPR1 promoter, facilitating its own expression. Intriguingly, CDK8 and its associated Mediator subunits positively regulate NPR1 and PR1 expression and play a pivotal role in local and systemic immunity. Moreover, CDK8 interacts with WRKY6, WRKY18, and TGA transcription factors and brings RNA polymerase II to NPR1 and PR1 promoters and coding regions to facilitate their expression. Our studies reveal a mechanism in which NPR1 recruits CDK8, WRKY18, and TGA transcription factors along with RNA polymerase II in the presence of SA and thereby facilitates its own and target gene expression for the establishment of plant immunity.
© 2019 American Society of Plant Biologists. All Rights Reserved.
Figures






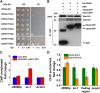
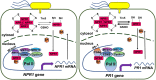
Comment in
-
NPR1 Has Everything under Control.Plant Physiol. 2019 Sep;181(1):6-7. doi: 10.1104/pp.19.00890. Plant Physiol. 2019. PMID: 31467138 Free PMC article. No abstract available.
References
-
- Axtell MJ, McNellis TW, Mudgett MB, Hsu CS, Staskawicz BJ (2001) Mutational analysis of the Arabidopsis RPS2 disease resistance gene and the corresponding Pseudomonas syringae avrRpt2 avirulence gene. Mol Plant Microbe Interact 14: 181–188 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

