Aβ34 is a BACE1-derived degradation intermediate associated with amyloid clearance and Alzheimer's disease progression
- PMID: 31110178
- PMCID: PMC6527709
- DOI: 10.1038/s41467-019-10152-w
Aβ34 is a BACE1-derived degradation intermediate associated with amyloid clearance and Alzheimer's disease progression
Abstract
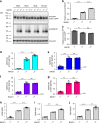
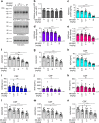
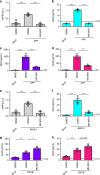
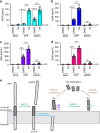
The beta-site APP cleaving enzyme 1 (BACE1) is known primarily for its initial cleavage of the amyloid precursor protein (APP), which ultimately leads to the generation of Aβ peptides. Here, we provide evidence that altered BACE1 levels and activity impact the degradation of Aβ40 and Aβ42 into a common Aβ34 intermediate. Using human cerebrospinal fluid (CSF) samples from the Amsterdam Dementia Cohort, we show that Aβ34 is elevated in individuals with mild cognitive impairment who later progressed to dementia. Furthermore, Aβ34 levels correlate with the overall Aβ clearance rates in amyloid positive individuals. Using CSF samples from the PREVENT-AD cohort (cognitively normal individuals at risk for Alzheimer's disease), we further demonstrate that the Aβ34/Aβ42 ratio, representing Aβ degradation and cortical deposition, associates with pre-clinical markers of neurodegeneration. We propose that Aβ34 represents a marker of amyloid clearance and may be helpful for the characterization of Aβ turnover in clinical samples.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

