IFN-γ-independent immune markers of Mycobacterium tuberculosis exposure
- PMID: 31110348
- PMCID: PMC6559862
- DOI: 10.1038/s41591-019-0441-3
IFN-γ-independent immune markers of Mycobacterium tuberculosis exposure
Erratum in
-
Publisher Correction: IFN-γ-independent immune markers of Mycobacterium tuberculosis exposure.Nat Med. 2019 Jul;25(7):1175. doi: 10.1038/s41591-019-0519-y. Nat Med. 2019. PMID: 31222179 Free PMC article.
Abstract
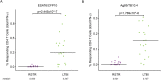
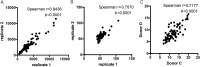
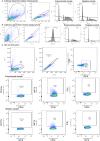
Exposure to Mycobacterium tuberculosis (Mtb) results in heterogeneous clinical outcomes including primary progressive tuberculosis and latent Mtb infection (LTBI). Mtb infection is identified using the tuberculin skin test and interferon-γ (IFN-γ) release assay IGRA, and a positive result may prompt chemoprophylaxis to prevent progression to tuberculosis. In the present study, we report on a cohort of Ugandan individuals who were household contacts of patients with TB. These individuals were highly exposed to Mtb but tested negative disease by IFN-γ release assay and tuberculin skin test, 'resisting' development of classic LTBI. We show that 'resisters' possess IgM, class-switched IgG antibody responses and non-IFN-γ T cell responses to the Mtb-specific proteins ESAT6 and CFP10, immunologic evidence of exposure to Mtb. Compared to subjects with classic LTBI, 'resisters' display enhanced antibody avidity and distinct Mtb-specific IgG Fc profiles. These data reveal a distinctive adaptive immune profile among Mtb-exposed subjects, supporting an expanded definition of the host response to Mtb exposure, with implications for public health and the design of clinical trials.
Conflict of interest statement
The authors declare no competing interests.
Figures












References
-
- World Health Organization. Global Tuberculosis Report 2018 (WHO, Geneva, 2018).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

