Mediterranean grassland soil C-N compound turnover is dependent on rainfall and depth, and is mediated by genomically divergent microorganisms
- PMID: 31110364
- PMCID: PMC6784897
- DOI: 10.1038/s41564-019-0449-y
Mediterranean grassland soil C-N compound turnover is dependent on rainfall and depth, and is mediated by genomically divergent microorganisms
Abstract
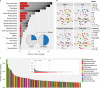
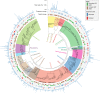
Soil microbial activity drives the carbon and nitrogen cycles and is an important determinant of atmospheric trace gas turnover, yet most soils are dominated by microorganisms with unknown metabolic capacities. Even Acidobacteria, among the most abundant bacteria in soil, remain poorly characterized, and functions across groups such as Verrucomicrobia, Gemmatimonadetes, Chloroflexi and Rokubacteria are understudied. Here, we have resolved 60 metagenomic and 20 proteomic data sets from a Mediterranean grassland soil ecosystem and recovered 793 near-complete microbial genomes from 18 phyla, representing around one-third of all microorganisms detected. Importantly, this enabled extensive genomics-based metabolic predictions for these communities. Acidobacteria from multiple previously unstudied classes have genomes that encode large enzyme complements for complex carbohydrate degradation. Alternatively, most microorganisms encode carbohydrate esterases that strip readily accessible methyl and acetyl groups from polymers like pectin and xylan, forming methanol and acetate, the availability of which could explain the high prevalence of C1 metabolism and acetate utilization in genomes. Microorganism abundances among samples collected at three soil depths and under natural and amended rainfall regimes indicate statistically higher associations of inorganic nitrogen metabolism and carbon degradation in deep and shallow soils, respectively. This partitioning decreased in samples under extended spring rainfall, indicating that long-term climate alteration can affect both carbon and nitrogen cycling. Overall, by leveraging natural and experimental gradients with genome-resolved metabolic profiles, we link microorganisms lacking prior genomic characterization to specific roles in complex carbon, C1, nitrate and ammonia transformations, and constrain factors that impact their distributions in soil.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Boval M, Dixon RM. The importance of grasslands for animal production and other functions: a review on management and methodological progress in the tropics. Animal. 2012;6:748–762. - PubMed
-
- Eze S, Palmer SM, Chapman PJ. Soil organic carbon stock in grasslands: effects of inorganic fertilizers, liming and grazing in different climate settings. J. Environ. Manage. 2018;223:74–84. - PubMed
-
- Delgado-Baquerizo M, et al. A global atlas of the dominant bacteria found in soil. Science. 2018;359:320–325. - PubMed

