Mycobacterial SigA and SigB Cotranscribe Essential Housekeeping Genes during Exponential Growth
- PMID: 31113892
- PMCID: PMC6529629
- DOI: 10.1128/mBio.00273-19
Mycobacterial SigA and SigB Cotranscribe Essential Housekeeping Genes during Exponential Growth
Abstract
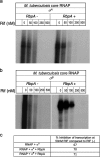
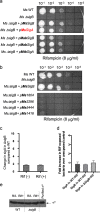
Mycobacterial σB belongs to the group II family of sigma factors, which are widely considered to transcribe genes required for stationary-phase survival and the response to stress. Here we explored the mechanism underlying the observed hypersensitivity of ΔsigB deletion mutants of Mycobacteriumsmegmatis, M. abscessus, and M. tuberculosis to rifampin (RIF) and uncovered an additional constitutive role of σB during exponential growth of mycobacteria that complements the function of the primary sigma factor, σA Using chromatin immunoprecipitation sequencing (ChIP-Seq), we show that during exponential phase, σB binds to over 200 promoter regions, including those driving expression of essential housekeeping genes, like the rRNA gene. ChIP-Seq of ectopically expressed σA-FLAG demonstrated that at least 61 promoter sites are recognized by both σA and σB These results together suggest that RNA polymerase holoenzymes containing either σA or σB transcribe housekeeping genes in exponentially growing mycobacteria. The RIF sensitivity of the ΔsigB mutant possibly reflects a decrease in the effective housekeeping holoenzyme pool, which results in susceptibility of the mutant to lower doses of RIF. Consistent with this model, overexpression of σA restores the RIF tolerance of the ΔsigB mutant to that of the wild type, concomitantly ruling out a specialized role of σB in RIF tolerance. Although the properties of mycobacterial σB parallel those of Escherichiacoli σ38 in its ability to transcribe a subset of housekeeping genes, σB presents a clear departure from the E. coli paradigm, wherein the cellular levels of σ38 are tightly controlled during exponential growth, such that the transcription of housekeeping genes is initiated exclusively by a holoenzyme containing σ70 (E.σ70).IMPORTANCE All mycobacteria encode a group II sigma factor, σB, closely related to the group I principal housekeeping sigma factor, σA Group II sigma factors are widely believed to play specialized roles in the general stress response and stationary-phase transition in the bacteria that encode them. Contrary to this widely accepted view, we show an additional housekeeping function of σB that complements the function of σA in logarithmically growing cells. These findings implicate a novel and dynamic partnership between σA and σB in maintaining the expression of housekeeping genes in mycobacteria and can perhaps be extended to other bacterial species that possess multiple group II sigma factors.
Keywords: ChIP-Seq; Mycobacterium; rifampin; sigB; sigma factor.
Copyright © 2019 Hurst-Hess et al.
Figures




Similar articles
-
Transcription of two sigma 70 homologue genes, sigA and sigB, in stationary-phase Mycobacterium tuberculosis.J Bacteriol. 1999 Jan;181(2):469-76. doi: 10.1128/JB.181.2.469-476.1999. J Bacteriol. 1999. PMID: 9882660 Free PMC article.
-
RbpA and σB association regulates polyphosphate levels to modulate mycobacterial isoniazid-tolerance.Mol Microbiol. 2018 Jun;108(6):627-640. doi: 10.1111/mmi.13952. Epub 2018 Apr 6. Mol Microbiol. 2018. PMID: 29575247
-
A mycobacterial extracytoplasmic sigma factor involved in survival following heat shock and oxidative stress.J Bacteriol. 1999 Jul;181(14):4266-74. doi: 10.1128/JB.181.14.4266-4274.1999. J Bacteriol. 1999. PMID: 10400584 Free PMC article.
-
Extra and intracellular expression of Mycobacterium tuberculosis genes.Tuber Lung Dis. 1998;79(2):91-7. doi: 10.1054/tuld.1998.0010. Tuber Lung Dis. 1998. PMID: 10645446 Review.
-
Sigma factors and promoters in Corynebacterium glutamicum.J Biotechnol. 2011 Jul 10;154(2-3):101-13. doi: 10.1016/j.jbiotec.2011.01.017. Epub 2011 Jan 26. J Biotechnol. 2011. PMID: 21277915 Review.
Cited by
-
M. tuberculosis Transcription Machinery: A Review on the Mycobacterial RNA Polymerase and Drug Discovery Efforts.Life (Basel). 2022 Nov 3;12(11):1774. doi: 10.3390/life12111774. Life (Basel). 2022. PMID: 36362929 Free PMC article. Review.
-
Mycobacterium tuberculosis Requires the Outer Membrane Lipid Phthiocerol Dimycocerosate for Starvation-Induced Antibiotic Tolerance.mSystems. 2023 Feb 23;8(1):e0069922. doi: 10.1128/msystems.00699-22. Epub 2023 Jan 4. mSystems. 2023. PMID: 36598240 Free PMC article.
-
The Rip1 intramembrane protease contributes to iron and zinc homeostasis in Mycobacterium tuberculosis.mSphere. 2023 Aug 24;8(4):e0038922. doi: 10.1128/msphere.00389-22. Epub 2023 Jun 15. mSphere. 2023. PMID: 37318217 Free PMC article.
-
In Vivo Methods to Study Protein-Protein Interactions as Key Players in Mycobacterium Tuberculosis Virulence.Pathogens. 2019 Oct 1;8(4):173. doi: 10.3390/pathogens8040173. Pathogens. 2019. PMID: 31581602 Free PMC article. Review.
-
Ethanol in Combination with Oxidative Stress Significantly Impacts Mycobacterial Physiology.J Bacteriol. 2020 Nov 4;202(23):e00222-20. doi: 10.1128/JB.00222-20. Print 2020 Nov 4. J Bacteriol. 2020. PMID: 32928928 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
