Transcriptional profiling of coaggregation interactions between Streptococcus gordonii and Veillonella parvula by Dual RNA-Seq
- PMID: 31113978
- PMCID: PMC6529473
- DOI: 10.1038/s41598-019-43979-w
Transcriptional profiling of coaggregation interactions between Streptococcus gordonii and Veillonella parvula by Dual RNA-Seq
Abstract
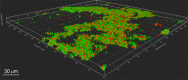
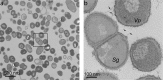
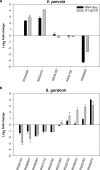
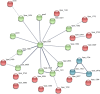
Many oral bacteria form macroscopic clumps known as coaggregates when mixed with a different species. It is thought that these cell-cell interactions are critical for the formation of mixed-species biofilms such as dental plaque. Here, we assessed the impact of coaggregation between two key initial colonizers of dental plaque, Streptococcus gordonii and Veillonella parvula, on gene expression in each partner. These species were shown to coaggregate in buffer or human saliva. To monitor gene regulation, coaggregates were formed in human saliva and, after 30 minutes, whole-transcriptomes were extracted for sequencing and Dual RNA-Seq analysis. In total, 272 genes were regulated in V. parvula, including 39 genes in oxidoreductase processes. In S. gordonii, there was a high degree of inter-sample variation. Nevertheless, 69 genes were identified as potentially regulated by coaggregation, including two phosphotransferase system transporters and several other genes involved in carbohydrate metabolism. Overall, these data indicate that responses of V. parvula to coaggregation with S. gordonii are dominated by oxidative stress-related processes, whereas S. gordonii responses are more focussed on carbohydrate metabolism. We hypothesize that these responses may reflect changes in the local microenvironment in biofilms when S. gordonii or V. parvula immigrate into the system.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

