Caver Web 1.0: identification of tunnels and channels in proteins and analysis of ligand transport
- PMID: 31114897
- PMCID: PMC6602463
- DOI: 10.1093/nar/gkz378
Caver Web 1.0: identification of tunnels and channels in proteins and analysis of ligand transport
Abstract
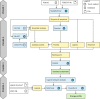
Caver Web 1.0 is a web server for comprehensive analysis of protein tunnels and channels, and study of the ligands' transport through these transport pathways. Caver Web is the first interactive tool allowing both the analyses within a single graphical user interface. The server is built on top of the abundantly used tunnel detection tool Caver 3.02 and CaverDock 1.0 enabling the study of the ligand transport. The program is easy-to-use as the only required inputs are a protein structure for a tunnel identification and a list of ligands for the transport analysis. The automated guidance procedures assist the users to set up the calculation in a way to obtain biologically relevant results. The identified tunnels, their properties, energy profiles and trajectories for ligands' passages can be calculated and visualized. The tool is very fast (2-20 min per job) and is applicable even for virtual screening purposes. Its simple setup and comprehensive graphical user interface make the tool accessible for a broad scientific community. The server is freely available at https://loschmidt.chemi.muni.cz/caverweb.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

Similar articles
-
Fully automated virtual screening pipeline of FDA-approved drugs using Caver Web.Comput Struct Biotechnol J. 2022 Nov 17;20:6512-6518. doi: 10.1016/j.csbj.2022.11.031. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36467577 Free PMC article.
-
Caver Web 2.0: analysis of tunnels and ligand transport in dynamic ensembles of proteins.Nucleic Acids Res. 2025 Jul 7;53(W1):W132-W142. doi: 10.1093/nar/gkaf399. Nucleic Acids Res. 2025. PMID: 40337920 Free PMC article.
-
CaverDock: a molecular docking-based tool to analyse ligand transport through protein tunnels and channels.Bioinformatics. 2019 Dec 1;35(23):4986-4993. doi: 10.1093/bioinformatics/btz386. Bioinformatics. 2019. PMID: 31077297
-
Fast approximative methods for study of ligand transport and rational design of improved enzymes for biotechnologies.Biotechnol Adv. 2022 Nov;60:108009. doi: 10.1016/j.biotechadv.2022.108009. Epub 2022 Jun 20. Biotechnol Adv. 2022. PMID: 35738509 Review.
-
Virtual screening of potential anticancer drugs based on microbial products.Semin Cancer Biol. 2022 Nov;86(Pt 2):1207-1217. doi: 10.1016/j.semcancer.2021.07.012. Epub 2021 Jul 21. Semin Cancer Biol. 2022. PMID: 34298109 Review.
Cited by
-
Simulation of Ligand Transport in Receptors Using CaverDock.Methods Mol Biol. 2021;2266:105-124. doi: 10.1007/978-1-0716-1209-5_6. Methods Mol Biol. 2021. PMID: 33759123
-
Characteristics of a new carotenoid cleavage dioxygenase NtCCD10 derived from Nicotiana tabacum.Planta. 2022 Oct 17;256(5):100. doi: 10.1007/s00425-022-04013-y. Planta. 2022. PMID: 36251100
-
Nucleotide binding to the ATP-cone in anaerobic ribonucleotide reductases allosterically regulates activity by modulating substrate binding.Elife. 2024 Jul 5;12:RP89292. doi: 10.7554/eLife.89292. Elife. 2024. PMID: 38968292 Free PMC article.
-
Structure-Based Design of Small Imine Reductase Panels for Target Substrates.ACS Catal. 2023 Sep 5;13(18):12310-12321. doi: 10.1021/acscatal.3c02278. eCollection 2023 Sep 15. ACS Catal. 2023. PMID: 37736118 Free PMC article.
-
CLICK-chemoproteomics and molecular dynamics simulation reveals pregnenolone targets and their binding conformations in Th2 cells.Front Immunol. 2023 Oct 31;14:1229703. doi: 10.3389/fimmu.2023.1229703. eCollection 2023. Front Immunol. 2023. PMID: 38022565 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

