Simple ClinVar: an interactive web server to explore and retrieve gene and disease variants aggregated in ClinVar database
- PMID: 31114901
- PMCID: PMC6602488
- DOI: 10.1093/nar/gkz411
Simple ClinVar: an interactive web server to explore and retrieve gene and disease variants aggregated in ClinVar database
Abstract
Clinical genetic testing has exponentially expanded in recent years, leading to an overwhelming amount of patient variants with high variability in pathogenicity and heterogeneous phenotypes. A large part of the variant level data is aggregated in public databases such as ClinVar. However, the ability to explore this rich resource and answer general questions such as 'How many genes inside ClinVar are associated with a specific disease? or 'In which part of the protein are patient variants located?' is limited and requires advanced bioinformatics processing. Here, we present Simple ClinVar (http://simple-clinvar.broadinstitute.org/) a web server application that is able to provide variant, gene and disease level summary statistics based on the entire ClinVar database in a dynamic and user-friendly web-interface. Overall, our web application is able to interactively answer basic questions regarding genetic variation and its known relationships to disease. By typing a disease term of interest, the user can identify in seconds the genes and phenotypes most frequently reported to ClinVar. Subsets of variants can then be further explored, filtered or mapped and visualized in the corresponding protein sequences. Our website will follow ClinVar monthly releases and provide easy access to ClinVar resources to a broader audience including basic and clinical scientists.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures

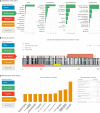

References
-
- O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D. et al. .. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–D745. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

