Direct role for the Drosophila GIGYF protein in 4EHP-mediated mRNA repression
- PMID: 31114929
- PMCID: PMC6648886
- DOI: 10.1093/nar/gkz429
Direct role for the Drosophila GIGYF protein in 4EHP-mediated mRNA repression
Abstract
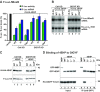
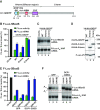
The eIF4E-homologous protein (4EHP) is a translational repressor that competes with eIF4E for binding to the 5'-cap structure of specific mRNAs, to which it is recruited by protein factors such as the GRB10-interacting GYF (glycine-tyrosine-phenylalanine domain) proteins (GIGYF). Several experimental evidences suggest that GIGYF proteins are not merely facilitating 4EHP recruitment to transcripts but are actually required for the repressor activity of the complex. However, the underlying molecular mechanism is unknown. Here, we investigated the role of the uncharacterized Drosophila melanogaster (Dm) GIGYF protein in post-transcriptional mRNA regulation. We show that, when in complex with 4EHP, Dm GIGYF not only elicits translational repression but also promotes target mRNA decay via the recruitment of additional effector proteins. We identified the RNA helicase Me31B/DDX6, the decapping activator HPat and the CCR4-NOT deadenylase complex as binding partners of GIGYF proteins. Recruitment of Me31B and HPat via discrete binding motifs conserved among metazoan GIGYF proteins is required for downregulation of mRNA expression by the 4EHP-GIGYF complex. Our findings are consistent with a model in which GIGYF proteins additionally recruit decapping and deadenylation complexes to 4EHP-containing RNPs to induce translational repression and degradation of mRNA targets.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



Similar articles
-
Molecular basis for GIGYF-Me31B complex assembly in 4EHP-mediated translational repression.Genes Dev. 2019 Oct 1;33(19-20):1355-1360. doi: 10.1101/gad.329219.119. Epub 2019 Aug 22. Genes Dev. 2019. PMID: 31439631 Free PMC article.
-
HPat provides a link between deadenylation and decapping in metazoa.J Cell Biol. 2010 Apr 19;189(2):289-302. doi: 10.1083/jcb.200910141. J Cell Biol. 2010. PMID: 20404111 Free PMC article.
-
GIGYF1/2 proteins use auxiliary sequences to selectively bind to 4EHP and repress target mRNA expression.Genes Dev. 2017 Jun 1;31(11):1147-1161. doi: 10.1101/gad.299420.117. Epub 2017 Jul 11. Genes Dev. 2017. PMID: 28698298 Free PMC article.
-
eIF4E-homologous protein (4EHP): a multifarious cap-binding protein.FEBS J. 2023 Jan;290(2):266-285. doi: 10.1111/febs.16275. Epub 2021 Nov 29. FEBS J. 2023. PMID: 34758096 Review.
-
Contrasting mechanisms of regulating translation of specific Drosophila germline mRNAs at the level of 5'-cap structure binding.Biochem Soc Trans. 2005 Dec;33(Pt 6):1544-6. doi: 10.1042/BST0331544. Biochem Soc Trans. 2005. PMID: 16246166 Review.
Cited by
-
The TUTase URT1 connects decapping activators and prevents the accumulation of excessively deadenylated mRNAs to avoid siRNA biogenesis.Nat Commun. 2021 Feb 26;12(1):1298. doi: 10.1038/s41467-021-21382-2. Nat Commun. 2021. PMID: 33637717 Free PMC article.
-
The EIF4E1-4EIP cap-binding complex of Trypanosoma brucei interacts with the terminal uridylyl transferase TUT3.PLoS One. 2021 Nov 22;16(11):e0258903. doi: 10.1371/journal.pone.0258903. eCollection 2021. PLoS One. 2021. PMID: 34807934 Free PMC article.
-
GIGYF2: A Multifunctional Regulator at the Crossroads of Gene Expression, mRNA Surveillance, and Human Disease.Cells. 2025 Jul 5;14(13):1032. doi: 10.3390/cells14131032. Cells. 2025. PMID: 40643551 Free PMC article. Review.
-
PARP inhibitor radiosensitization enhances anti-PD-L1 immunotherapy through stabilizing chemokine mRNA in small cell lung cancer.Nat Commun. 2025 Mar 4;16(1):2166. doi: 10.1038/s41467-025-57257-z. Nat Commun. 2025. PMID: 40038278 Free PMC article.
-
Translational repression of NMD targets by GIGYF2 and EIF4E2.PLoS Genet. 2021 Oct 19;17(10):e1009813. doi: 10.1371/journal.pgen.1009813. eCollection 2021 Oct. PLoS Genet. 2021. PMID: 34665823 Free PMC article.
References
-
- Rom E., Kim H.C., Gingras A.C., Marcotrigiano J., Favre D., Olsen H., Burley S.K., Sonenberg N.. Cloning and characterization of 4EHP, a novel mammalian eIF4E-related cap-binding protein. J. Biol. Chem. 1998; 273:13104–13109. - PubMed
-
- Joshi B., Cameron A., Jagus R.. Characterization of mammalian eIF4E-family members. Eur. J. Biochem. 2004; 271:2189–2203. - PubMed
-
- Cho P.F., Poulin F., Cho-Park Y.A., Cho-Park I.B., Chicoine J.D., Lasko P., Sonenberg N.. A new paradigm for translational control: inhibition via 5′-3′ mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell. 2005; 121:411–423. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

