Integrated bioinformatic analysis of differentially expressed genes and signaling pathways in plaque psoriasis
- PMID: 31115544
- PMCID: PMC6580009
- DOI: 10.3892/mmr.2019.10241
Integrated bioinformatic analysis of differentially expressed genes and signaling pathways in plaque psoriasis
Abstract
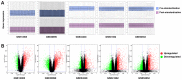
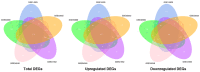
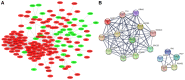
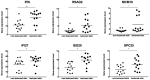
Psoriasis is an immune‑mediated cutaneous disorder with a high incidence and prevalence. Patients with psoriasis may experience irritation, pain and psychological problems. The cause and underlying molecular etiology of psoriasis remains unknown. In an attempt to achieve a more comprehensive understanding of the molecular pathogenesis of psoriasis, the gene expression profiles of 175 pairs of lesional and corresponding non‑lesional skin samples were downloaded from 5 data sets in the Gene Expression Omnibus (GEO) database. Integrated differentially expressed genes (DEGs) were obtained with the use of R software. The gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment were analyzed using the DAVID online analysis tool. The protein‑protein interaction (PPI) network was constructed on the STRING platform and hub genes were calculated with the use of Cytoscape software. Finally, GEO2R was used to determine the expression of the hub genes in scalp psoriasis. A total of 373 genes from the 5 data sets were identified as DEGs, including 277 upregulated and 96 downregulated genes. GO analysis revealed that immune responses and epidermal differentiation/development were the most enriched terms in biological processes, extracellular space/matrix was the most enriched term in cellular components, and endopeptidase inhibitor activity was the most enriched term in molecular functions. In the KEGG pathway enrichment, DEGs were mainly enriched in the metabolic and viral infection‑associated pathways. A total of 17 hub genes were calculated, including CSK2, CDC45, MCM10, SPC25, NDC80, NUF2, AURKA, CENPE, RRM2, DLGP5, HMMR, TTK, IFIT1, RSAD2, IFI6, IFI27 and ISG20, among which interferon‑α‑inducible genes were revealed to display a similar expression pattern as that obtained in scalp psoriasis. This comprehensive bioinformatic re‑analysis of GEO data provides new insights on the molecular pathogenesis of psoriasis and the identification of potential therapeutic targets for the treatment of psoriasis.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

