Screening key lncRNAs for human rectal adenocarcinoma based on lncRNA-mRNA functional synergistic network
- PMID: 31116002
- PMCID: PMC6639256
- DOI: 10.1002/cam4.2236
Screening key lncRNAs for human rectal adenocarcinoma based on lncRNA-mRNA functional synergistic network
Abstract
Background: Rectal adenocarcinoma (READ) is one of the deadliest malignancies, and the molecular mechanisms underlying the initiation and development of READ remain largely unknown. In this study, we aimed to find key long noncoding RNAs (lncRNAs) and mRNAs in READ by RNA sequencing.
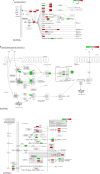
Methods: RNA sequencing was performed to identify differentially expressed mRNAs (DEmRNAs) and lncRNAs (DElncRNAs) between READ and normal tissue. READ-specific protein-protein interaction (PPI), DElncRNA-DEmRNA coexpression, and DElncRNA-nearby DEmRNA interaction networks were constructed. DEmRNAs and DEmRNAs coexpressed with DElncRNAs were functionally annotated.
Results: A total of 2113 DEmRNAs and 150 DElncRNAs between READ and normal tissue were identified. The PPI network identified several hub proteins, including CDK1, AURKB, CDC6, FOXQ1, NUF2, and TOP2A. The DElncRNA-DEmRNA coexpression and DElncRNA-nearby DEmRNA interaction networks identified some hub lncRNAs, including CCAT1, LOC105374879, GAS5, and B3GALT5-AS1. The colorectal cancer pathway, the intestinal immune network for IgA production and the p53 signaling pathway were three pathways significantly enriched in DEmRNAs and DEmRNAs coexpressed with DElncRNAs. MSH6 coexpressed with two DElncRNAs (LOC105374879 and CASC15) and BCL2 coexpressed with B3GALT5-AS1 were significantly enriched in the colorectal cancer signaling pathway. TNFRSF17 coexpressed with B3GALT5-AS1 was enriched in the intestinal immune network for IgA production. CCNB2 coexpressed with LOC105374879 was enriched in the p53 signaling pathway.
Conclusion: A total of four DEmRNAs (MSH6, BCL2, TNFRSF17, and CCNB2) and three DElncRNAs (LOC105374879, CASC15, and B3GALT5-AS1) may be involved in the pathogenesis of READ; this data may contribute to understanding the mechanisms of READ and the development of therapeutic strategies for READ.
Keywords: DElncRNA-DEmRNA coexpression; DElncRNAs; DEmRNAs; RNA sequencing; rectal adenocarcinoma.
© 2019 The Authors. Cancer Medicine published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no conflict of interest. No competing financial interests exist.
Figures






Similar articles
-
Identification of aberrantly expressed long non-coding RNAs in postmenopausal osteoporosis.Int J Mol Med. 2018 Jun;41(6):3537-3550. doi: 10.3892/ijmm.2018.3575. Epub 2018 Mar 19. Int J Mol Med. 2018. PMID: 29568943 Free PMC article.
-
Identification of aberrantly expressed long non-coding RNAs in stomach adenocarcinoma.Oncotarget. 2017 Jul 25;8(30):49201-49216. doi: 10.18632/oncotarget.17329. Oncotarget. 2017. PMID: 28484081 Free PMC article.
-
Screening of tumor grade-related mRNAs and lncRNAs for Esophagus Squamous Cell Carcinoma.J Clin Lab Anal. 2021 Jun;35(6):e23797. doi: 10.1002/jcla.23797. Epub 2021 May 7. J Clin Lab Anal. 2021. PMID: 33960436 Free PMC article.
-
Identification of MFI2-AS1, a Novel Pivotal lncRNA for Prognosis of Stage III/IV Colorectal Cancer.Dig Dis Sci. 2020 Dec;65(12):3538-3550. doi: 10.1007/s10620-020-06064-1. Epub 2020 Jan 20. Dig Dis Sci. 2020. PMID: 31960204 Review.
-
Reconstruction and Analysis of the Differentially Expressed IncRNA-miRNA-mRNA Network Based on Competitive Endogenous RNA in Hepatocellular Carcinoma.Crit Rev Eukaryot Gene Expr. 2019;29(6):539-549. doi: 10.1615/CritRevEukaryotGeneExpr.2019028740. Crit Rev Eukaryot Gene Expr. 2019. PMID: 32422009 Review.
Cited by
-
Integrative Analysis of Regulatory Module Reveals Associations of Microgravity with Dysfunctions of Multi-body Systems and Tumorigenesis.Int J Mol Sci. 2020 Oct 14;21(20):7585. doi: 10.3390/ijms21207585. Int J Mol Sci. 2020. PMID: 33066530 Free PMC article.
-
Construction of a clinical survival prognostic model for middle-aged and elderly patients with stage III rectal adenocarcinoma.World J Clin Cases. 2021 Mar 6;9(7):1563-1579. doi: 10.12998/wjcc.v9.i7.1563. World J Clin Cases. 2021. PMID: 33728300 Free PMC article.
-
Microarray analysis of differentially expressed long non-coding RNAs in daidzein-treated lung cancer cells.Oncol Lett. 2021 Nov;22(5):789. doi: 10.3892/ol.2021.13050. Epub 2021 Sep 17. Oncol Lett. 2021. PMID: 34630702 Free PMC article.
-
Identification of Core Genes Involved in the Metastasis of Clear Cell Renal Cell Carcinoma.Cancer Manag Res. 2020 Dec 30;12:13437-13449. doi: 10.2147/CMAR.S276818. eCollection 2020. Cancer Manag Res. 2020. PMID: 33408516 Free PMC article.
-
Uncovering potential genes in colorectal cancer based on integrated and DNA methylation analysis in the gene expression omnibus database.BMC Cancer. 2022 Feb 3;22(1):138. doi: 10.1186/s12885-022-09185-0. BMC Cancer. 2022. PMID: 35114976 Free PMC article.
References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics. CA Cancer J Clin. 2016;66(2016):7‐30. - PubMed
-
- Torre LA, Bray F, Siegel RL, Ferlay J, Lortet‐Tieulent J, Jemal A. Global cancer statistics. CA Cancer J Clin. 2012;65(2015):87‐108. - PubMed
-
- Li Y, Shi X, Yang W, et al. Transcriptome profiling of lncRNA and co‐expression networks in esophageal squamous cell carcinoma by RNA sequencing. Tumour Biol. 2016;37:13091‐13100. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

