AlphaFold at CASP13
- PMID: 31116374
- PMCID: PMC6907002
- DOI: 10.1093/bioinformatics/btz422
AlphaFold at CASP13
Abstract
Computational prediction of protein structure from sequence is broadly viewed as a foundational problem of biochemistry and one of the most difficult challenges in bioinformatics. Once every two years the Critical Assessment of protein Structure Prediction (CASP) experiments are held to assess the state of the art in the field in a blind fashion, by presenting predictor groups with protein sequences whose structures have been solved but have not yet been made publicly available. The first CASP was organized in 1994, and the latest, CASP13, took place last December, when for the first time the industrial laboratory DeepMind entered the competition. DeepMind's entry, AlphaFold, placed first in the Free Modeling (FM) category, which assesses methods on their ability to predict novel protein folds (the Zhang group placed first in the Template-Based Modeling (TBM) category, which assess methods on predicting proteins whose folds are related to ones already in the Protein Data Bank.) DeepMind's success generated significant public interest. Their approach builds on two ideas developed in the academic community during the preceding decade: (i) the use of co-evolutionary analysis to map residue co-variation in protein sequence to physical contact in protein structure, and (ii) the application of deep neural networks to robustly identify patterns in protein sequence and co-evolutionary couplings and convert them into contact maps. In this Letter, we contextualize the significance of DeepMind's entry within the broader history of CASP, relate AlphaFold's methodological advances to prior work, and speculate on the future of this important problem.
© The Author(s) 2019. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures
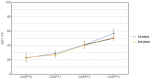
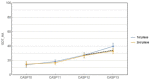
References
-
- Anand N. et al. (2018) Generative modeling for protein structures In: Bengio S.et al. (eds) Advances in Neural Information Processing Systems 31. Curran Associates, Inc, Montreal, pp. 7505–7516.
-
- Bepler T., Berger B. (2018) Learning protein sequence embeddings using information from structure. ICLR 2019.
-
- Brünger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr, 54, 905–921. - PubMed

