ER-phagy: shaping up and destressing the endoplasmic reticulum
- PMID: 31116513
- PMCID: PMC6772018
- DOI: 10.1111/febs.14932
ER-phagy: shaping up and destressing the endoplasmic reticulum
Abstract
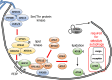
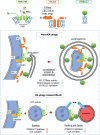
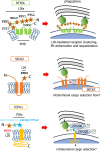
The endoplasmic reticulum (ER) network has central roles in metabolism and cellular organization. The ER undergoes dynamic alterations in morphology, molecular composition and functional specification. Remodelling of the network under fluctuating conditions enables the continual performance of ER functions and minimizes stress. Recent data have revealed that selective autophagy-mediated degradation of ER fragments, or ER-phagy, fundamentally contributes to this remodelling. This review provides a perspective on established views of selective autophagy, comparing these with emerging mechanisms of ER-phagy and related processes. The text discusses the impact of ER-phagy on the function of the ER- and the cell, both in normal physiology and when dysregulated within disease settings. Finally, unanswered questions regarding the mechanisms and significance of ER-phagy are highlighted.
Keywords: ERLAD; ER-phagy; FAM134B; microautophagy; recovER-phagy.
© 2019 The Authors. The FEBS Journal published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Walter P & Ron D (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

