Circuit diversification in a biofilm regulatory network
- PMID: 31116789
- PMCID: PMC6530872
- DOI: 10.1371/journal.ppat.1007787
Circuit diversification in a biofilm regulatory network
Erratum in
-
Correction: Circuit diversification in a biofilm regulatory network.PLoS Pathog. 2019 Jul 19;15(7):e1007966. doi: 10.1371/journal.ppat.1007966. eCollection 2019 Jul. PLoS Pathog. 2019. PMID: 31323072 Free PMC article.
Abstract
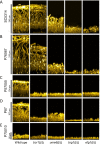
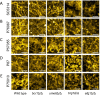
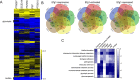
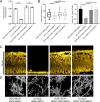
Genotype-phenotype relationships can vary extensively among members of a species. One cause of this variation is circuit diversification, the alteration of gene regulatory relationships among members of a species. Circuit diversification is thought to be a starting point for the circuit divergence or rewiring that occurs during speciation. How widespread is circuit diversification? Here we address this question with the fungal pathogen Candida albicans, which forms biofilms rich in distinctive hyphal cells as a prelude to infection. Our understanding of the biofilm/hyphal regulatory network comes primarily from studies of one clinical isolate, strain SC5314, and its marked derivatives. We used CRISPR-based methods to create mutations of four key biofilm transcription factor genes-BCR1, UME6, BRG1, and EFG1 -in SC5314 and four additional clinical isolates. Phenotypic analysis revealed that mutations in BCR1 or UME6 have variable impact across strains, while mutations in BRG1 or EFG1 had uniformly severe impact. Gene expression, sampled with Nanostring probes and examined comprehensively for EFG1 via RNA-Seq, indicates that regulatory relationships are highly variable among isolates. Our results suggest that genotype-phenotype relationships vary in this strain panel in part because of differences in control of BRG1 by BCR1, a hypothesis that is supported through engineered constitutive expression of BRG1. Overall, the data show that circuit diversification is the rule, not the exception, in this biofilm/hyphal regulatory network.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

