The lncRNA SLNCR Recruits the Androgen Receptor to EGR1-Bound Genes in Melanoma and Inhibits Expression of Tumor Suppressor p21
- PMID: 31116991
- PMCID: PMC6668037
- DOI: 10.1016/j.celrep.2019.04.101
The lncRNA SLNCR Recruits the Androgen Receptor to EGR1-Bound Genes in Melanoma and Inhibits Expression of Tumor Suppressor p21
Abstract
Melanoma is the deadliest form of skin cancer, affecting men more frequently and severely than women. Although recent studies suggest that differences in activity of the androgen receptor (AR) underlie the observed sex bias, little is known about AR activity in melanoma. Here we show that AR and EGR1 bind to the long non-coding RNA SLNCR and increase melanoma proliferation through coordinated transcriptional regulation of several growth-regulatory genes. ChIP-seq reveals that ligand-free AR is enriched on SLNCR-regulated melanoma genes and that AR genomic occupancy significantly overlaps with EGR1 at consensus EGR1 binding sites. We present a model in which SLNCR recruits AR to EGR1-bound genomic loci and switches EGR1-mediated transcriptional activation to repression of the tumor suppressor p21Waf1/Cip1. Our data implicate the regulatory triad of SLNCR, AR, and EGR1 in promoting oncogenesis and may help explain why men have a higher incidence of and more rapidly progressive melanomas compared with women.
Keywords: CDKN1A; EGR1; Waf1/Cip1; androgen receptor; linc00673; long non-coding RNA; melanoma; metastasis; p21; proliferation.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
K.S. and E.Y. are current employees of Alnylam Pharmaceuticals. J.C. is a current employee of the University of Toronto. D.D.T. is a current employee of Rubius Therapeutics. K.S. and C.N. are inventors of two patents based on work presented in this manuscript (PCT/US2016/041343 and PCT/US2018/050597).
Figures

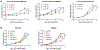


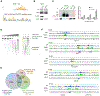
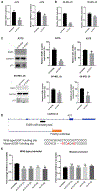

References
-
- Ahmed MM, Venkatasubbarao K, Fruitwala SM, Muthukkumar S, Wood DP Jr., Sells SF, Mohiuddin M, and Rangnekar VM (1996). EGR-1 induction is required for maximal radiosensitivity in A375-C6 melanoma cells. J. Biol. Chem. 277, 29231–29237. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

