Exploring the Evolution of Virulence Factors through Bioinformatic Data Mining
- PMID: 31117023
- PMCID: PMC6529551
- DOI: 10.1128/mSystems.00162-19
Exploring the Evolution of Virulence Factors through Bioinformatic Data Mining
Abstract
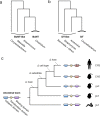
The molecular evolution of virulence factors is a central theme in our understanding of bacterial pathogenesis and host-microbe interactions. Using bioinformatics and genome data mining, recent studies have shed light on the evolution of important virulence factor families and the mechanisms by which they have adapted and diversified in function. This perspective highlights three complementary approaches useful for studying the molecular evolution of virulence factors: identification and analysis of virulence factor homologs, detection of adaptations or functional shifts, and computational prediction of novel virulence factor families. Each of these research directions is associated with distinct questions, approaches, and challenges for future work. Moving forward, bioinformatics will continue to play a critical role in exploring the evolution of virulence factors, including those that target humans. By reconstructing past processes and events, we will be able to better interpret newly sequenced microbial genomes and detect future pathoadaptations.
Keywords: bioinformatics; microbial genomics; molecular evolution; pathogens; virulence factors.
Copyright © 2019 Doxey et al.
Conflict of interest statement
Conflict of Interest Disclosures: A.C.D. has nothing to disclose. M.J.M. has nothing to disclose. B.L. has nothing to disclose.
Figures
References
LinkOut - more resources
Full Text Sources
Miscellaneous

