Mechanism of CK2.3, a Novel Mimetic Peptide of Bone Morphogenetic Protein Receptor Type IA, Mediated Osteogenesis
- PMID: 31117181
- PMCID: PMC6567251
- DOI: 10.3390/ijms20102500
Mechanism of CK2.3, a Novel Mimetic Peptide of Bone Morphogenetic Protein Receptor Type IA, Mediated Osteogenesis
Abstract
Background: Osteoporosis is a degenerative skeletal disease with a limited number of treatment options. CK2.3, a novel peptide, may be a potential therapeutic. It induces osteogenesis and bone formation in vitro and in vivo by acting downstream of BMPRIA through releasing CK2 from the receptor. However, the detailed signaling pathways, the time frame of signaling, and genes activated remain largely unknown.
Methods: Using a newly developed fluorescent CK2.3 analog, specific inhibitors for the BMP signaling pathways, Western blot, and RT-qPCR, we determined the mechanism of CK2.3 in C2C12 cells. We then confirmed the results in primary BMSCs.
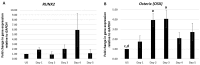
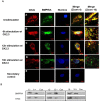
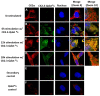
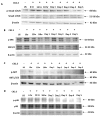
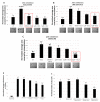
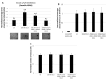
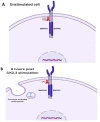
Results: Using these methods, we showed that CK2.3 stimulation activated OSX, ALP, and OCN. CK2.3 stimulation induced time dependent release of CK2β from BMPRIA and concurrently CK2.3 colocalized with CK2α. Furthermore, CK2.3 induced BMP signaling depends on ERK1/2 and Smad1/5/8 signaling pathways.
Conclusion: CK2.3 is a novel peptide that drives osteogenesis, and we detailed the molecular sequence of events that are triggered from the stimulation of CK2.3 until the induction of mineralization. This knowledge can be applied in the development of future therapeutics for osteoporosis.
Keywords: BMP2; CK2; CK2.3; RT-qPCR; bone; osteogenesis; osteogenic signal transduction; osteoporosis; western blots.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Raisz L.G. Physiology and pathophysiology of bone remodeling. Clin. Chem. 1999;45:1353–1358. - PubMed
MeSH terms
Substances
Grants and funding
- R01 AR064243/AR/NIAMS NIH HHS/United States
- 1R01AR064242-010A1/National Institute of Arthritis and Musculoskeletal and Skin Diseases of the National Institutes of Health
- P20GM103446/Institutional Development Award (IDeA) from the National Institute of Health's National Institute of General Medical Sciences
LinkOut - more resources
Full Text Sources
Miscellaneous

