Level of neo-epitope predecessor and mutation type determine T cell activation of MHC binding peptides
- PMID: 31118084
- PMCID: PMC6532181
- DOI: 10.1186/s40425-019-0595-z
Level of neo-epitope predecessor and mutation type determine T cell activation of MHC binding peptides
Abstract
Background: Targeting epitopes derived from neo-antigens (or "neo-epitopes") represents a promising immunotherapy approach with limited off-target effects. However, most peptides predicted using MHC binding prediction algorithms do not induce a CD8 + T cell response, and there is a crucial need to refine the predictions to readily identify the best antigens that could mediate T-cell responses. Such a response requires a high enough number of epitopes bound to the target MHC. This number is correlated with both the peptide-MHC binding affinity and the number of peptides reaching the ER. Beyond this, the response may be affected by the properties of the neo-epitope mutated residues.
Methods: Herein, we analyzed several experimental datasets from cancer patients to elaborate better predictive algorithms for T-cell reactivity to neo-epitopes.
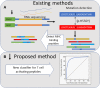
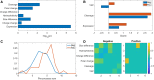
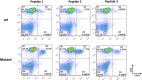
Results: Indeed, potent classifiers for epitopes derived from neo-antigens in melanoma and other tumors can be developed based on biochemical properties of the mutated residue, the antigen expression level and the peptide processing stage. Among MHC binding peptides, the present classifiers can remove half of the peptides falsely predicted to activate T cells while maintaining the absolute majority of reactive peptides.
Conclusions: The classifier properties further highlight the contribution of the quantity of peptides reaching the ER and the mutation type to CD8 + T cell responses. These classifiers were then validated on neo-antigens obtained from other datasets, confirming the validity of our prediction. Algorithm Availability: http://peptibase.cs.biu.ac.il/Tcell_predictor/ or by request from the authors as a standalone code.
Keywords: MHC-binding peptides; Machine Learning; Neoantigen; T cell activation.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
MuPeXI: prediction of neo-epitopes from tumor sequencing data.Cancer Immunol Immunother. 2017 Sep;66(9):1123-1130. doi: 10.1007/s00262-017-2001-3. Epub 2017 Apr 20. Cancer Immunol Immunother. 2017. PMID: 28429069 Free PMC article.
-
Benchmarking predictions of MHC class I restricted T cell epitopes in a comprehensively studied model system.PLoS Comput Biol. 2020 May 26;16(5):e1007757. doi: 10.1371/journal.pcbi.1007757. eCollection 2020 May. PLoS Comput Biol. 2020. PMID: 32453790 Free PMC article.
-
Somatically mutated regions of immunoglobulin on human B-cell lymphomas code for peptides that bind to autologous major histocompatibility complex class I, providing a potential target for cytotoxic T cells.Cancer Res. 2001 Jul 1;61(13):5145-52. Cancer Res. 2001. PMID: 11431353
-
Recent Advances in Lung Cancer Immunotherapy: Input of T-Cell Epitopes Associated With Impaired Peptide Processing.Front Immunol. 2019 Jul 3;10:1505. doi: 10.3389/fimmu.2019.01505. eCollection 2019. Front Immunol. 2019. PMID: 31333652 Free PMC article. Review.
-
New treatment options for patients with melanoma: review of melanoma-derived T-cell epitope-based peptide vaccines.Melanoma Res. 1996 Feb;6(1):11-24. doi: 10.1097/00008390-199602000-00003. Melanoma Res. 1996. PMID: 8640065 Review.
Cited by
-
The Current Landscape of mRNA Vaccines Against Viruses and Cancer-A Mini Review.Front Immunol. 2022 May 6;13:885371. doi: 10.3389/fimmu.2022.885371. eCollection 2022. Front Immunol. 2022. PMID: 35603213 Free PMC article. Review.
-
Cancer Vaccines: Antigen Selection Strategy.Vaccines (Basel). 2021 Jan 25;9(2):85. doi: 10.3390/vaccines9020085. Vaccines (Basel). 2021. PMID: 33503926 Free PMC article. Review.
-
Naive and memory T cells TCR-HLA-binding prediction.Oxf Open Immunol. 2022 May 26;3(1):iqac001. doi: 10.1093/oxfimm/iqac001. eCollection 2022. Oxf Open Immunol. 2022. PMID: 36846560 Free PMC article.
-
Informing immunotherapy with multi-omics driven machine learning.NPJ Digit Med. 2024 Mar 14;7(1):67. doi: 10.1038/s41746-024-01043-6. NPJ Digit Med. 2024. PMID: 38486092 Free PMC article. Review.
-
A domain-based vaccine construct against SARS-CoV-2, the causative agent of COVID-19 pandemic: development of self-amplifying mRNA and peptide vaccines.Bioimpacts. 2021;11(1):65-84. doi: 10.34172/bi.2021.11. Epub 2020 Dec 10. Bioimpacts. 2021. PMID: 33469510 Free PMC article.
References
-
- Yewdell JW, Reits E, Neefjes J. Making sense of mass destruction: quantitating MHC class I antigen presentation. Nat Rev Immunol [Internet]. 2003 [cited 2018 Jan 30];3(12):952–961. Available from: http://www.nature.com/doifinder/10.1038/nri1250 - DOI - PubMed
-
- Singh SP, Mishra BN. Major histocompatibility complex linked databases and prediction tools for designing vaccines. Hum Immunol [Internet]. 2016 [cited 2017 Nov 29];77(3):295–306. Available from: http://www.sciencedirect.com/science/article/pii/S0198885915005650 - PubMed
-
- Luo H, Ye H, Ng HW, Shi L, Tong W, Mendrick DL, et al. Machine Learning Methods for Predicting HLA-Peptide Binding Activity. Bioinform Biol Insights [Internet]. 2015 [cited 2017 Nov 29];9(Suppl 3):21–29. Available from: https://journals.sagepub.com/doi/pdf/10.4137/BBI.S29466. - DOI - PMC - PubMed
-
- Tran E, Robbins PF, Rosenberg SA. “Final common pathway” of human cancer immunotherapy: targeting random somatic mutations. Nat Immunol [Internet]. 2017 [cited 2017 Nov 29];18(3):255–262. Available from: http://www.nature.com/doifinder/10.1038/ni.3682 - DOI - PMC - PubMed
-
- Sharma P, Allison JP. The future of immune checkpoint therapy. Science [Internet]. 2015 [cited 2017 Nov 20];348(6230):56–61. Available from: https://science.sciencemag.org/content/348/6230/56.long. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
