Junín Virus Promotes Autophagy To Facilitate the Virus Life Cycle
- PMID: 31118257
- PMCID: PMC6639288
- DOI: 10.1128/JVI.02307-18
Junín Virus Promotes Autophagy To Facilitate the Virus Life Cycle
Abstract
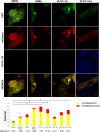
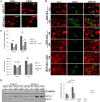
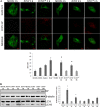
Junín virus (JUNV), a member of the family Arenaviridae, is the etiological agent of Argentine hemorrhagic fever (AHF), a potentially deadly endemic-epidemic disease affecting the population of the most fertile farming land of Argentina. Autophagy is a degradative process with a crucial antiviral role; however, several viruses subvert the pathway to their benefit. We determined the role of autophagy in JUNV-infected cells by analyzing LC3, a cytoplasmic protein (LC3-I) that becomes vesicle membrane associated (LC3-II) upon induction of autophagy. Cells overexpressing enhanced green fluorescent protein (EGFP)-LC3 and infected with JUNV showed an increased number of LC3 punctate structures, similar to those obtained after starvation or bafilomycin A1 treatment, which leads to autophagosome induction or accumulation, respectively. We also monitored the conversion of LC3-I to LC3-II, observing LC3-II levels in JUNV-infected cells similar to those observed in starved cells. Additionally, we kinetically studied the number of LC3 dots after JUNV infection and found that the virus activated the pathway as early as 2 h postinfection (p.i.), whereas the UV-inactivated virus did not induce the pathway. Cells subjected to starvation or pretreated with rapamycin, a pharmacological autophagy inductor, enhanced virus yield. Also, we assayed the replication capacity of JUNV in Atg5 knockout or Beclin 1 knockdown cells (both critical components of the autophagic pathway) and found a significant decrease in JUNV replication. Taken together, our results constitute the first study indicating that JUNV infection induces an autophagic response, which is functionally required by the virus for efficient propagation.IMPORTANCE Mammalian arenaviruses are zoonotic viruses that cause asymptomatic and persistent infections in their rodent hosts but may produce severe and lethal hemorrhagic fevers in humans. Currently, there are neither effective therapeutic options nor effective vaccines for viral hemorrhagic fevers caused by human-pathogenic arenaviruses, except the vaccine Candid no. 1 against Argentine hemorrhagic fever (AHF), licensed for human use in areas of endemicity in Argentina. Since arenaviruses remain a severe threat to global public health, more in-depth knowledge of their replication mechanisms would improve our ability to fight these viruses. Autophagy is a lysosomal degradative pathway involved in maintaining cellular homeostasis, representing powerful anti-infective machinery. We show, for the first time for a member of the family Arenaviridae, a proviral role of autophagy in JUNV infection, providing new knowledge in the field of host-virus interaction. Therefore, modulation of virus-induced autophagy could be used as a strategy to block arenavirus infections.
Keywords: Arenaviridae; Beclin 1; Junín virus; autophagy; innate immunity; proviral role; replication.
Copyright © 2019 American Society for Microbiology.
Figures


References
-
- Buchmeier MJ. 2007. Arenaviridae: the virus and their replication, p 1792–1827. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields Virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA.
-
- Briese T, Paweska JT, McMullan LK, Hutchison SK, Street C, Palacios G, Khristova ML, Weyer J, Swanepoel R, Egholm M, Nichol ST, Lipkin WI. 2009. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever–associated arenavirus from Southern Africa. PLoS Pathog 5:e1000455. doi:10.1371/journal.ppat.1000455. - DOI - PMC - PubMed
-
- Parodi AS, Greenway DJ, Rugiero HR, Frigerio M, De La Barrera JM, Mettler N, Garzon F, Boxaca M, Guerrero L, Nota N. 1958. Concerning the epidemic outbreak in Junin. Dia Med 30:2300–2301. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

