Pre-analytic Considerations for Mass Spectrometry-Based Untargeted Metabolomics Data
- PMID: 31119672
- PMCID: PMC7346099
- DOI: 10.1007/978-1-4939-9236-2_20
Pre-analytic Considerations for Mass Spectrometry-Based Untargeted Metabolomics Data
Abstract
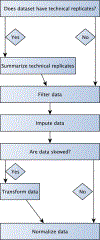
Metabolomics is the science of characterizing and quantifying small molecule metabolites in biological systems. These metabolites give organisms their biochemical characteristics, providing a link between genotype, environment, and phenotype. With these opportunities also come data challenges, such as compound annotation, missing values, and batch effects. We present the steps of a general pipeline to process untargeted mass spectrometry data to alleviate the latter two challenges. We assume to have a matrix with metabolite abundances, with metabolites in rows and samples in columns. The steps in the pipeline include summarizing technical replicates (if available), filtering, imputing, transforming, and normalizing the data. In each of these steps, a method and parameters should be chosen based on assumptions one is willing to make, the question of interest, and diagnostic tools. Besides giving a general pipeline that can be adapted by the reader, our goal is to review diagnostic tools and criteria that are helpful when making decisions in each step of the pipeline and assessing the effectiveness of normalization and batch correction. We conclude by giving a list of useful packages and discuss some alternative approaches that might be more appropriate for the reader's data.
Keywords: Filtering; Imputation; Mass spectrometry; Metabolomics; Normalization; Pre-analytic; Processing; Technical replicates; Untargeted.
Figures
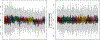
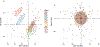
References
-
- Jordan KW, Nordenstam J, Lauwers GY, Rothenberger DA, Alavi K, Garwood M, Cheng LL. Metabolomic Characterization of Human Rectal Adenocarcinoma with Intact Tissue Magnetic Resonance Spectroscopy. Diseases of the Colon & Rectum. 2009;52(3):520–5. doi: 10.1007/DCR.0b013e31819c9a2c. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

