An Arabidopsis FANCJ helicase homologue is required for DNA crosslink repair and rDNA repeat stability
- PMID: 31120885
- PMCID: PMC6550410
- DOI: 10.1371/journal.pgen.1008174
An Arabidopsis FANCJ helicase homologue is required for DNA crosslink repair and rDNA repeat stability
Abstract
Proteins of the Fanconi Anemia (FA) complementation group are required for crosslink (CL) repair in humans and their loss leads to severe pathological phenotypes. Here we characterize a homolog of the Fe-S cluster helicase FANCJ in the model plant Arabidopsis, AtFANCJB, and show that it is involved in interstrand CL repair. It acts at a presumably early step in concert with the nuclease FAN1 but independently of the nuclease AtMUS81, and is epistatic to both error-prone and error-free post-replicative repair in Arabidopsis. The simultaneous knock out of FANCJB and the Fe-S cluster helicase RTEL1 leads to induced cell death in root meristems, indicating an important role of the enzymes in replicative DNA repair. Surprisingly, we found that AtFANCJB is involved in safeguarding rDNA stability in plants. In the absence of AtRTEL1 and AtFANCJB, we detected a synergetic reduction to about one third of the original number of 45S rDNA copies. It is tempting to speculate that the detected rDNA instability might be due to deficiencies in G-quadruplex structure resolution and might thus contribute to pathological phenotypes of certain human genetic diseases.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
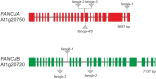




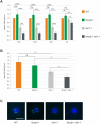

References
-
- Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, et al. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001; 105: 149–160. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

