Exome-wide search and functional annotation of genes associated in patients with severe tick-borne encephalitis in a Russian population
- PMID: 31122248
- PMCID: PMC6533173
- DOI: 10.1186/s12920-019-0503-x
Exome-wide search and functional annotation of genes associated in patients with severe tick-borne encephalitis in a Russian population
Abstract
Background: Tick-borne encephalitis (TBE) is a viral infectious disease caused by tick-borne encephalitis virus (TBEV). TBEV infection is responsible for a variety of clinical manifestations ranging from mild fever to severe neurological illness. Genetic factors involved in the host response to TBEV that may potentially play a role in the severity of the disease are still poorly understood. In this study, using whole-exome sequencing, we aimed to identify genetic variants and genes associated with severe forms of TBE as well as biological pathways through which the identified variants may influence the severity of the disease.
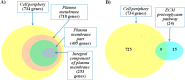
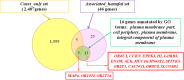
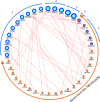
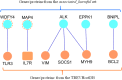
Results: Whole-exome sequencing data analysis was performed on 22 Russian patients with severe forms of TBE and 17 Russian individuals from the control group. We identified 2407 candidate genes harboring rare, potentially pathogenic variants in exomes of patients with TBE and not containing any rare, potentially pathogenic variants in exomes of individuals from the control group. According to DAVID tool, this set of 2407 genes was enriched with genes involved in extracellular matrix proteoglycans pathway and genes encoding proteins located at the cell periphery. A total of 154 genes/proteins from these functional groups have been shown to be involved in protein-protein interactions (PPIs) with the known candidate genes/proteins extracted from TBEVHostDB database. By ranking these genes according to the number of rare harmful minor alleles, we identified two genes (MSR1 and LMO7), harboring five minor alleles, and three genes (FLNA, PALLD, PKD1) harboring four minor alleles. When considering genes harboring genetic variants associated with severe forms of TBE at the suggestive P-value < 0.01, 46 genes containing harmful variants were identified. Out of these 46 genes, eight (MAP4, WDFY4, ACTRT2, KLHL25, MAP2K3, MBD1, OR10J1, and OR2T34) were additionally found among genes containing rare pathogenic variants identified in patients with TBE; and five genes (WDFY4, ALK, MAP4, BNIPL, EPPK1) were found to encode proteins that are involved in PPIs with proteins encoded by genes from TBEVHostDB. Three genes out of five (MAP4, EPPK1, ALK) were found to encode proteins located at cell periphery.
Conclusions: Whole-exome sequencing followed by systems biology approach enabled to identify eight candidate genes (MAP4, WDFY4, ACTRT2, KLHL25, MAP2K3, MBD1, OR10J1, and OR2T34) that can potentially determine predisposition to severe forms of TBE. Analyses of the genetic risk factors for severe forms of TBE revealed a significant enrichment with genes controlling extracellular matrix proteoglycans pathway as well as genes encoding components of cell periphery.
Keywords: Biological pathways; Candidate genes; Flavivirus; Genetic predisposition; Network; PPIs; Tick-borne encephalitis; Whole-exome sequencing.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures
Similar articles
-
A matrix metalloproteinase 9 (MMP9) gene single nucleotide polymorphism is associated with predisposition to tick-borne encephalitis virus-induced severe central nervous system disease.Ticks Tick Borne Dis. 2018 May;9(4):763-767. doi: 10.1016/j.ttbdis.2018.02.010. Epub 2018 Feb 17. Ticks Tick Borne Dis. 2018. PMID: 29496490
-
A database of human genes and a gene network involved in response to tick-borne encephalitis virus infection.BMC Evol Biol. 2017 Dec 28;17(Suppl 2):259. doi: 10.1186/s12862-017-1107-8. BMC Evol Biol. 2017. PMID: 29297316 Free PMC article.
-
Tick-borne encephalitis in Europe and Russia: Review of pathogenesis, clinical features, therapy, and vaccines.Antiviral Res. 2019 Apr;164:23-51. doi: 10.1016/j.antiviral.2019.01.014. Epub 2019 Jan 31. Antiviral Res. 2019. PMID: 30710567 Review.
-
Association of single nucleotide polymorphism rs3775291 in the coding region of the TLR3 gene with predisposition to tick-borne encephalitis in a Russian population.Antiviral Res. 2013 Aug;99(2):136-8. doi: 10.1016/j.antiviral.2013.05.008. Epub 2013 May 27. Antiviral Res. 2013. PMID: 23721942
-
EAN consensus review on prevention, diagnosis and management of tick-borne encephalitis.Eur J Neurol. 2017 Oct;24(10):1214-e61. doi: 10.1111/ene.13356. Epub 2017 Aug 1. Eur J Neurol. 2017. PMID: 28762591 Review.
Cited by
-
Genotype-driven sensitivity of mice to tick-borne encephalitis virus correlates with differential host responses in peripheral macrophages and brain.J Neuroinflammation. 2025 Jan 28;22(1):22. doi: 10.1186/s12974-025-03354-1. J Neuroinflammation. 2025. PMID: 39875898 Free PMC article.
-
Whole genome sequencing reveals population diversity and variation in HIV-1 specific host genes.Front Genet. 2023 Dec 20;14:1290624. doi: 10.3389/fgene.2023.1290624. eCollection 2023. Front Genet. 2023. PMID: 38179408 Free PMC article.
-
Computer genomics research at the bioinformatics conference series in Novosibirsk.BMC Genomics. 2019 Jul 11;20(Suppl 7):537. doi: 10.1186/s12864-019-5846-3. BMC Genomics. 2019. PMID: 31291908 Free PMC article. No abstract available.
-
Human plasmacytoid dendritic cells at the crossroad of type I interferon-regulated B cell differentiation and antiviral response to tick-borne encephalitis virus.PLoS Pathog. 2021 Apr 15;17(4):e1009505. doi: 10.1371/journal.ppat.1009505. eCollection 2021 Apr. PLoS Pathog. 2021. PMID: 33857267 Free PMC article.
-
Whole-Exome Sequencing Analysis of Human Semen Quality in Russian Multiethnic Population.Front Genet. 2021 Jun 11;12:662846. doi: 10.3389/fgene.2021.662846. eCollection 2021. Front Genet. 2021. PMID: 34178030 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

