miR-301a promotes lung tumorigenesis by suppressing Runx3
- PMID: 31122259
- PMCID: PMC6532219
- DOI: 10.1186/s12943-019-1024-0
miR-301a promotes lung tumorigenesis by suppressing Runx3
Abstract
Background: Our previous report demonstrated that genetic ablation of miR-301a reduces Kras-driven lung tumorigenesis in mice. However, the impact of miR-301a on host anti-tumor immunity remains unexplored. Here we assessed the underlying molecular mechanisms of miR-301a in the tumor microenvironment.
Methods: The differentially expressed genes were identified by using deep sequencing. The immune cell counts, and cytokines expression were analyzed by realtime PCR, immunohistochemistry and flow cytometry. The role of miR-301a/Runx3 in lung tumor was evaluated on cell growth, migration and invasion. The function of miR-301a/Runx3 in regulating tumor microenvironment and tumor metastasis were evaluated in Kras transgenic mice and B16/LLC1 syngeneic xenografts tumor models.
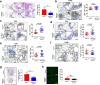
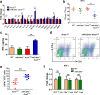
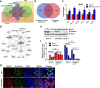
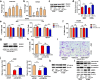
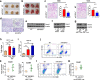
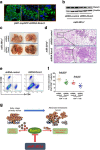
Results: In this work, we identified 1166 up-regulated and 475 down-regulated differentially expressed genes in lung tumor tissues between KrasLA2 and miR-301a-/-; KrasLA2 mice. Immune response and cell cycle were major pathways involved in the protective role of miR-301a deletion in lung tumorigenesis. Overexpression of the miR-301a target, Runx3, was an early event identified in miR-301a-/-; KrasLA2 mice compared to WT-KrasLA2 mice. We found that miR-301a deletion enhanced CD8+ T cell accumulation and IFN-γ production in the tumor microenvironment and mediated antitumor immunity. Further studies revealed that miR-301a deficiency in the tumor microenvironment effectively reduced tumor metastasis by elevating Runx3 and recruiting CD8+ T cells, whereas miR-301a knockdown in tumor cells themselves restrained cell migration by elevating Runx3 expression.
Conclusions: Our findings further underscore that miR-301a facilitates tumor microenvironment antitumor immunity by Runx3 suppression in lung tumorigenesis.
Keywords: CD8+ T cells; IFN-γ; Kras; Runx3; miR-301a.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Overexpression of miR-301a-3p promotes colorectal cancer cell proliferation and metastasis by targeting deleted in liver cancer-1 and runt-related transcription factor 3.J Cell Biochem. 2019 Apr;120(4):6078-6089. doi: 10.1002/jcb.27894. Epub 2018 Oct 25. J Cell Biochem. 2019. PMID: 30362160
-
Overexpressed miR-301a promotes cell proliferation and invasion by targeting RUNX3 in gastric cancer.J Gastroenterol. 2013 Sep;48(9):1023-33. doi: 10.1007/s00535-012-0733-6. Epub 2013 Jan 22. J Gastroenterol. 2013. PMID: 23338485
-
Downregulation of lncRNA MIR17HG reduced tumorigenicity and Treg-mediated immune escape of non-small-cell lung cancer cells through targeting the miR-17-5p/RUNX3 axis.J Biochem Mol Toxicol. 2024 May;38(5):e23715. doi: 10.1002/jbt.23715. J Biochem Mol Toxicol. 2024. PMID: 38704830
-
Multifaceted role of microRNA-301a in human cancer: from biomarker potential to therapeutic targeting.Cancer Gene Ther. 2024 Dec;31(12):1754-1764. doi: 10.1038/s41417-024-00832-1. Epub 2024 Sep 24. Cancer Gene Ther. 2024. PMID: 39317714 Review.
-
Molecular mechanisms of microRNA-301a during tumor progression and metastasis.Pathol Res Pract. 2023 Jul;247:154538. doi: 10.1016/j.prp.2023.154538. Epub 2023 May 18. Pathol Res Pract. 2023. PMID: 37209575 Review.
Cited by
-
RUNX3 is up-regulated in abdominal aortic aneurysm and regulates the function of vascular smooth muscle cells by regulating TGF-β1.J Mol Histol. 2022 Feb;53(1):1-11. doi: 10.1007/s10735-021-10035-9. Epub 2021 Nov 23. J Mol Histol. 2022. PMID: 34813022
-
HIF-1ɑ-regulated miR-1275 maintains stem cell-like phenotypes and promotes the progression of LUAD by simultaneously activating Wnt/β-catenin and Notch signaling.Theranostics. 2020 Jan 22;10(6):2553-2570. doi: 10.7150/thno.41120. eCollection 2020. Theranostics. 2020. PMID: 32194819 Free PMC article.
-
MicroRNA-301a-3p promotes triple-negative breast cancer progression through downregulating MEOX2.Exp Ther Med. 2021 Sep;22(3):945. doi: 10.3892/etm.2021.10377. Epub 2021 Jul 1. Exp Ther Med. 2021. PMID: 34306209 Free PMC article.
-
Immunogenomic features of radiologically distinctive nodules in multiple primary lung cancer.Cancer Immunol Immunother. 2024 Sep 5;73(11):217. doi: 10.1007/s00262-024-03807-1. Cancer Immunol Immunother. 2024. PMID: 39235522 Free PMC article.
-
Anesthetics may modulate cancer surgical outcome: a possible role of miRNAs regulation.BMC Anesthesiol. 2021 Mar 9;21(1):71. doi: 10.1186/s12871-021-01294-w. BMC Anesthesiol. 2021. PMID: 33750303 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

