MITF-the first 25 years
- PMID: 31123060
- PMCID: PMC6672050
- DOI: 10.1101/gad.324657.119
MITF-the first 25 years
Abstract
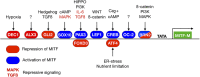
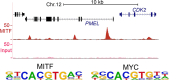
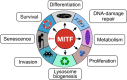
All transcription factors are equal, but some are more equal than others. In the 25 yr since the gene encoding the microphthalmia-associated transcription factor (MITF) was first isolated, MITF has emerged as a key coordinator of many aspects of melanocyte and melanoma biology. Like all transcription factors, MITF binds to specific DNA sequences and up-regulates or down-regulates its target genes. What marks MITF as being remarkable among its peers is the sheer range of biological processes that it appears to coordinate. These include cell survival, differentiation, proliferation, invasion, senescence, metabolism, and DNA damage repair. In this article we present our current understanding of MITF's role and regulation in development and disease, as well as those of the MITF-related factors TFEB and TFE3, and highlight key areas where our knowledge of MITF regulation and function is limited.
Keywords: MITF; MiT family; melanocytes; melanoma; transcription factor.
© 2019 Goding and Arnheiter; Published by Cold Spring Harbor Laboratory Press.
Figures








References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
