Recompleting the Caenorhabditis elegans genome
- PMID: 31123080
- PMCID: PMC6581061
- DOI: 10.1101/gr.244830.118
Recompleting the Caenorhabditis elegans genome
Abstract
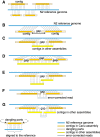
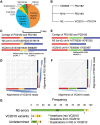
Caenorhabditis elegans was the first multicellular eukaryotic genome sequenced to apparent completion. Although this assembly employed a standard C. elegans strain (N2), it used sequence data from several laboratories, with DNA propagated in bacteria and yeast. Thus, the N2 assembly has many differences from any C. elegans available today. To provide a more accurate C. elegans genome, we performed long-read assembly of VC2010, a modern strain derived from N2. Our VC2010 assembly has 99.98% identity to N2 but with an additional 1.8 Mb including tandem repeat expansions and genome duplications. For 116 structural discrepancies between N2 and VC2010, 97 structures matching VC2010 (84%) were also found in two outgroup strains, implying deficiencies in N2. Over 98% of N2 genes encoded unchanged products in VC2010; moreover, we predicted ≥53 new genes in VC2010. The recompleted genome of C. elegans should be a valuable resource for genetics, genomics, and systems biology.
© 2019 Yoshimura et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


Comment in
-
A new reference genome sequence for Caenorhabditis elegans?Lab Anim (NY). 2019 Sep;48(9):267-268. doi: 10.1038/s41684-019-0371-1. Lab Anim (NY). 2019. PMID: 31406358 Free PMC article. No abstract available.
Similar articles
-
CGC1, a new reference genome for Caenorhabditis elegans.Genome Res. 2025 Aug 1;35(8):1902-1918. doi: 10.1101/gr.280274.124. Genome Res. 2025. PMID: 40664475 Free PMC article.
-
Genomic sequence of a mutant strain of Caenorhabditis elegans with an altered recombination pattern.BMC Genomics. 2010 Feb 23;11:131. doi: 10.1186/1471-2164-11-131. BMC Genomics. 2010. PMID: 20178641 Free PMC article.
-
Long-read sequencing reveals intra-species tolerance of substantial structural variations and new subtelomere formation in C. elegans.Genome Res. 2019 Jun;29(6):1023-1035. doi: 10.1101/gr.246082.118. Epub 2019 May 23. Genome Res. 2019. PMID: 31123081 Free PMC article.
-
Comparative genomics in C. elegans, C. briggsae, and other Caenorhabditis species.Methods Mol Biol. 2006;351:13-29. doi: 10.1385/1-59745-151-7:13. Methods Mol Biol. 2006. PMID: 16988423 Review.
-
Genomics in C. elegans: so many genes, such a little worm.Genome Res. 2005 Dec;15(12):1651-60. doi: 10.1101/gr.3729105. Genome Res. 2005. PMID: 16339362 Review.
Cited by
-
Units containing telomeric repeats are prevalent in subtelomeric regions of a Mesorhabditis isolate collected from the Republic of Korea.Genes Genomics. 2024 Dec;46(12):1461-1472. doi: 10.1007/s13258-024-01576-w. Epub 2024 Oct 4. Genes Genomics. 2024. PMID: 39367283
-
Sensory neurons couple arousal and foraging decisions in Caenorhabditis elegans.Elife. 2023 Dec 27;12:RP88657. doi: 10.7554/eLife.88657. Elife. 2023. PMID: 38149996 Free PMC article.
-
GALA: a computational framework for de novo chromosome-by-chromosome assembly with long reads.Nat Commun. 2023 Jan 13;14(1):204. doi: 10.1038/s41467-022-35670-y. Nat Commun. 2023. PMID: 36639368 Free PMC article.
-
A ubiquinone precursor analogue does not clearly increase the growth rate of Caenorhabditis inopinata.MicroPubl Biol. 2024 Dec 5;2024:10.17912/micropub.biology.001235. doi: 10.17912/micropub.biology.001235. eCollection 2024. MicroPubl Biol. 2024. PMID: 39712935 Free PMC article.
-
Expansion of the split hygromycin toolkit for transgene insertion in Caenorhabditis elegans.MicroPubl Biol. 2024 Jan 29;2024:10.17912/micropub.biology.001091. doi: 10.17912/micropub.biology.001091. eCollection 2024. MicroPubl Biol. 2024. PMID: 38351905 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
