Tumor suppression of novel anti-PD-1 antibodies mediated through CD28 costimulatory pathway
- PMID: 31123083
- PMCID: PMC6605749
- DOI: 10.1084/jem.20182359
Tumor suppression of novel anti-PD-1 antibodies mediated through CD28 costimulatory pathway
Abstract
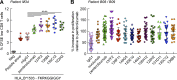
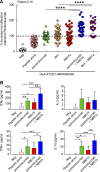
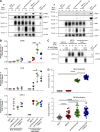
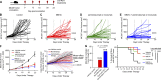
Classical antagonistic antibodies (Abs) targeting PD-1, such as pembrolizumab and nivolumab, act through blockade of the PD-1-PDL-1 interaction. Here, we have identified novel antagonistic anti-PD-1 Abs not blocking the PD-1-PDL-1 interaction. The nonblocking Abs recognize epitopes on PD-1 located on the opposing face of the PDL-1 interaction and overlap with a newly identified evolutionarily conserved patch. These nonblocking Abs act predominantly through the CD28 coreceptor. Importantly, a combination of blocking and nonblocking Abs synergize in the functional recovery of antigen-specific exhausted CD8 T cells. Interestingly, nonblocking anti-PD-1 Abs have equivalent antitumor activity compared with blocker Abs in two mouse tumor models, and combination therapy using both classes of Abs enhanced tumor suppression in the mouse immunogenic tumor model. The identification of the novel nonblocker anti-PD-1 Abs and their synergy with classical blocker Abs may be instrumental in potentiating immunotherapy strategies and antitumor activity.
© 2019 Fenwick et al.
Figures


References
-
- Adams, P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. . 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221. 10.1107/S0907444909052925 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

