Structural determinants for peptide-bond formation by asparaginyl ligases
- PMID: 31123145
- PMCID: PMC6576118
- DOI: 10.1073/pnas.1818568116
Structural determinants for peptide-bond formation by asparaginyl ligases
Abstract
Asparaginyl endopeptidases (AEPs) are cysteine proteases which break Asx (Asn/Asp)-Xaa bonds in acidic conditions. Despite sharing a conserved overall structure with AEPs, certain plant enzymes such as butelase 1 act as a peptide asparaginyl ligase (PAL) and catalyze Asx-Xaa bond formation in near-neutral conditions. PALs also serve as macrocyclases in the biosynthesis of cyclic peptides. Here, we address the question of how a PAL can function as a ligase rather than a protease. Based on sequence homology of butelase 1, we identified AEPs and PALs from the cyclic peptide-producing plants Viola yedoensis (Vy) and Viola canadensis (Vc) of the Violaceae family. Using a crystal structure of a PAL obtained at 2.4-Å resolution coupled to mutagenesis studies, we discovered ligase-activity determinants flanking the S1 site, namely LAD1 and LAD2 located around the S2 and S1' sites, respectively, which modulate ligase activity by controlling the accessibility of water or amine nucleophile to the S-ester intermediate. Recombinantly expressed VyPAL1-3, predicted to be PALs, were confirmed to be ligases by functional studies. In addition, mutagenesis studies on VyPAL1-3, VyAEP1, and VcAEP supported our prediction that LAD1 and LAD2 are important for ligase activity. In particular, mutagenesis targeting LAD2 selectively enhanced the ligase activity of VyPAL3 and converted the protease VcAEP into a ligase. The definition of structural determinants required for ligation activity of the asparaginyl ligases presented here will facilitate genomic identification of PALs and engineering of AEPs into PALs.
Keywords: data mining; ligase-activity determinant; peptide ligase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
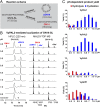



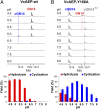
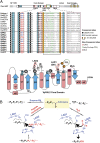
Similar articles
-
Site-Specific Protein Modifications by an Engineered Asparaginyl Endopeptidase from Viola canadensis.Front Chem. 2021 Oct 22;9:768854. doi: 10.3389/fchem.2021.768854. eCollection 2021. Front Chem. 2021. PMID: 34746098 Free PMC article.
-
Substrate-binding glycine residues are major determinants for hydrolase and ligase activity of plant legumains.New Phytol. 2023 May;238(4):1534-1545. doi: 10.1111/nph.18841. Epub 2023 Mar 14. New Phytol. 2023. PMID: 36843268
-
Molecular basis for the production of cyclic peptides by plant asparaginyl endopeptidases.Nat Commun. 2018 Jun 20;9(1):2411. doi: 10.1038/s41467-018-04669-9. Nat Commun. 2018. PMID: 29925835 Free PMC article.
-
Make it or break it: Plant AEPs on stage in biotechnology.Biotechnol Adv. 2020 Dec;45:107651. doi: 10.1016/j.biotechadv.2020.107651. Epub 2020 Oct 23. Biotechnol Adv. 2020. PMID: 33141031 Review.
-
Macrocyclization by asparaginyl endopeptidases.New Phytol. 2018 May;218(3):923-928. doi: 10.1111/nph.14511. Epub 2017 Mar 21. New Phytol. 2018. PMID: 28322452 Review.
Cited by
-
Approaches for peptide and protein cyclisation.Org Biomol Chem. 2021 May 12;19(18):3983-4001. doi: 10.1039/d1ob00411e. Org Biomol Chem. 2021. PMID: 33978044 Free PMC article. Review.
-
Asparaginyl Ligases with Engineered Substrate Specificity for Controlled, Sequential Transpeptidation Reactions.J Am Chem Soc. 2025 Jun 25;147(25):21824-21831. doi: 10.1021/jacs.5c04693. Epub 2025 Jun 11. J Am Chem Soc. 2025. PMID: 40499909 Free PMC article.
-
Chemoenzymatic Semisynthesis of Proteins.Chem Rev. 2020 Mar 25;120(6):3051-3126. doi: 10.1021/acs.chemrev.9b00450. Epub 2019 Nov 27. Chem Rev. 2020. PMID: 31774265 Free PMC article. Review.
-
An efficient and easily obtainable butelase variant for chemoenzymatic ligation and modification of peptides and proteins.Microb Cell Fact. 2024 Nov 30;23(1):325. doi: 10.1186/s12934-024-02598-5. Microb Cell Fact. 2024. PMID: 39614317 Free PMC article.
-
Vacuolar Processing Enzymes in Plant Programmed Cell Death and Autophagy.Int J Mol Sci. 2023 Jan 7;24(2):1198. doi: 10.3390/ijms24021198. Int J Mol Sci. 2023. PMID: 36674706 Free PMC article. Review.
References
-
- Mazmanian S. K., Liu G., Ton-That H., Schneewind O., Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285, 760–763 (1999). - PubMed
-
- Mao H., Hart S. A., Schink A., Pollok B. A., Sortase-mediated protein ligation: A new method for protein engineering. J. Am. Chem. Soc. 126, 2670–2671 (2004). - PubMed
-
- Piotukh K., et al. , Directed evolution of sortase A mutants with altered substrate selectivity profiles. J. Am. Chem. Soc. 133, 17536–17539 (2011). - PubMed
-
- Toplak A., Nuijens T., Quaedflieg P. J. L. M., Wu B., Janssen D. B., Peptiligase, an enzyme for efficient chemoenzymatic peptide synthesis and cyclization in water. Adv. Synth. Catal. 358, 2140–2147 (2016).
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

