Abnormal gut microbiota composition contributes to cognitive dysfunction in streptozotocin-induced diabetic mice
- PMID: 31123221
- PMCID: PMC6555457
- DOI: 10.18632/aging.101978
Abnormal gut microbiota composition contributes to cognitive dysfunction in streptozotocin-induced diabetic mice
Abstract
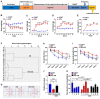
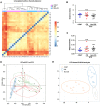
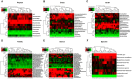
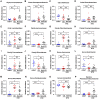
Both diabetes and Alzheimer's disease are age-related disorders, and numerous studies have demonstrated that patients with diabetes are at an increased risk of cognitive dysfunction (CD) and Alzheimer's disease, suggesting shared or interacting pathomechanisms. The present study investigated the role of abnormal gut microbiota in diabetes-induced CD and the potential underlying mechanisms. An intraperitoneal injection of streptozotocin administered for 5 consecutive days was used for establishing a diabetic animal model. Hierarchical cluster analysis of Morris water maze (MWM) performance indices (escape latency and target quadrant crossing) was adopted to classify the diabetic model mice into CD and Non-CD phenotypes. Both β-diversity and relative abundance of several gut bacteria significantly differed between the CD and Non-CD groups. Further, fecal bacteria transplantation from Non-CD mice, but not from CD mice, into the gut of pseudo-germ-free mice significantly improved host MWM performance, an effect associated with alterations in β-diversity and relative abundance of host gut bacteria. Collectively, these findings suggest that abnormal gut microbiota composition contributes to the onset of diabetes-induced CD and that improving gut microbiota composition is a potential therapeutic strategy for diabetes and related comorbidities.
Keywords: cognitive dysfunction; diabetes; gut microbiota; hierarchical cluster analysis; streptozotocin.
Conflict of interest statement
Figures


Similar articles
-
Reshaping the gut microbiota: Tangliping decoction and its core blood-absorbed component quercetin improve diabetic cognitive impairment.Phytomedicine. 2025 May;140:156560. doi: 10.1016/j.phymed.2025.156560. Epub 2025 Mar 5. Phytomedicine. 2025. PMID: 40058319
-
Abnormal gut microbiota composition contributes to cognitive dysfunction in SAMP8 mice.Aging (Albany NY). 2018 Jun 10;10(6):1257-1267. doi: 10.18632/aging.101464. Aging (Albany NY). 2018. PMID: 29886457 Free PMC article.
-
Abnormal gut microbiota composition is associated with experimental autoimmune prostatitis-induced depressive-like behaviors in mice.Prostate. 2020 Jun;80(9):663-673. doi: 10.1002/pros.23978. Epub 2020 Apr 7. Prostate. 2020. PMID: 32255522
-
The Gut Microbiota and Alzheimer's Disease.J Alzheimers Dis. 2017;58(1):1-15. doi: 10.3233/JAD-161141. J Alzheimers Dis. 2017. PMID: 28372330 Review.
-
Role of Gut Microbiota in Type 2 Diabetes Mellitus and Its Complications: Novel Insights and Potential Intervention Strategies.Korean J Gastroenterol. 2019 Dec 25;74(6):314-320. doi: 10.4166/kjg.2019.74.6.314. Korean J Gastroenterol. 2019. PMID: 31870137 Review.
Cited by
-
Heat Stress-Induced Dysbiosis of Porcine Colon Microbiota Plays a Role in Intestinal Damage: A Fecal Microbiota Profile.Front Vet Sci. 2022 Mar 1;9:686902. doi: 10.3389/fvets.2022.686902. eCollection 2022. Front Vet Sci. 2022. PMID: 35300220 Free PMC article.
-
Zi Shen Wan Fang Attenuates Neuroinflammation and Cognitive Function Via Remodeling the Gut Microbiota in Diabetes-Induced Cognitive Impairment Mice.Front Pharmacol. 2022 Jul 15;13:898360. doi: 10.3389/fphar.2022.898360. eCollection 2022. Front Pharmacol. 2022. PMID: 35910371 Free PMC article.
-
Gut Microbial Metabolite Short-Chain Fatt Acids Partially Reverse Surgery and Anesthesia-Induced Behavior Deficits in C57BL/6J Mice.Front Neurosci. 2021 Jun 8;15:664641. doi: 10.3389/fnins.2021.664641. eCollection 2021. Front Neurosci. 2021. PMID: 34168535 Free PMC article.
-
Fecal Microbiota Transplantation: A Microbiome Modulation Technique for Alzheimer's Disease.Cureus. 2021 Jul 20;13(7):e16503. doi: 10.7759/cureus.16503. eCollection 2021 Jul. Cureus. 2021. PMID: 34430117 Free PMC article. Review.
-
The Role of Gut Microbiota in Chronic Itch-Evoked Novel Object Recognition-Related Cognitive Dysfunction in Mice.Front Med (Lausanne). 2021 Feb 5;8:616489. doi: 10.3389/fmed.2021.616489. eCollection 2021. Front Med (Lausanne). 2021. PMID: 33614682 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

