Seed targeted RNAi-mediated silencing of GmMIPS1 limits phytate accumulation and improves mineral bioavailability in soybean
- PMID: 31123331
- PMCID: PMC6533290
- DOI: 10.1038/s41598-019-44255-7
Seed targeted RNAi-mediated silencing of GmMIPS1 limits phytate accumulation and improves mineral bioavailability in soybean
Abstract
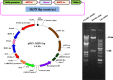
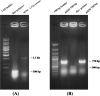
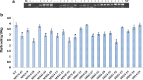
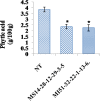
Phytic acid (PA), the major phosphorus reserve in soybean seeds (60-80%), is a potent ion chelator, causing deficiencies that leads to malnutrition. Several forward and reverse genetics approaches have ever since been explored to reduce its phytate levels to improve the micronutrient and phosphorous availability. Transgenic technology has met with success by suppressing the expression of the PA biosynthesis-related genes in several crops for manipulating their phytate content. In our study, we targeted the disruption of the expression of myo-inositol-3-phosphate synthase (MIPS1), the first and the rate limiting enzyme in PA biosynthesis in soybean seeds, by both antisense (AS) and RNAi approaches, using a seed specific promoter, vicilin. PCR and Southern analysis revealed stable integration of transgene in the advanced progenies. The transgenic seeds (T4) of AS (MS14-28-12-29-3-5) and RNAi (MI51-32-22-1-13-6) soybean lines showed 38.75% and 41.34% reduction in phytate levels respectively, compared to non-transgenic (NT) controls without compromised growth and seed development. The electron microscopic examination also revealed reduced globoid crystals in the Protein storage vacoules (PSVs) of mature T4 seeds compared to NT seed controls. A significant increase in the contents of Fe2+ (15.4%, 21.7%), Zn2+ (7.45%, 11.15%) and Ca2+ (10.4%, 15.35%) were observed in MS14-28-12-29-3-5 and MI51-32-22-1-13-6 transgenic lines, respectively, compared to NT implicating improved mineral bioavailability. This study signifies proof-of-concept demonstration of seed-specific PA reduction and paves the path towards low phytate soybean through pathway engineering using the new and precise editing tools.
Conflict of interest statement
The authors declare no competing interests.
Figures








References
-
- Raboy V, Dickinson DB, Below FE. Variation in seed total phosphorus, phytic acid, zinc, calcium, magnesium, and protein among lines of Glycine max and G. soja. Crop Sci. 1984;24:431–434.
-
- Kumar A, Kumar V, Lal SK, Jolly M, Sachdev A. Influence of gamma rays and ethyl methane sulphonate (EMS) on the levels of phytic acid, raffinose family oligosaccharides and antioxidants in soybean seeds of different genotypes. J. Plant Biochem. Biotechnol. 2015;24:204–209.
-
- Wilcox JR, Premachandra GS, Young KA, Raboy V. Isolation of high seed inorganic P, low phytate soybean mutants. Crop Sci. 2000;40:1601–1605.
-
- Zhou JR, Erdman JW., Jr. Phytic acid in health and disease. Crit. Rev. Food. Sci. Nutr. 1995;35:495–508. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

