Personalized medicine-concepts, technologies, and applications in inflammatory skin diseases
- PMID: 31124204
- PMCID: PMC6851586
- DOI: 10.1111/apm.12934
Personalized medicine-concepts, technologies, and applications in inflammatory skin diseases
Abstract
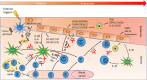
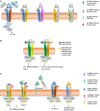
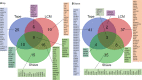
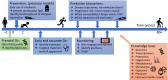
The current state, tools, and applications of personalized medicine with special emphasis on inflammatory skin diseases like psoriasis and atopic dermatitis are discussed. Inflammatory pathways are outlined as well as potential targets for monoclonal antibodies and small-molecule inhibitors.
Keywords: Atopic dermatitis; endotypes; immunology; inflammatory skin diseases; personalized medicine; precision medicine; psoriasis; targeted therapy.
© 2019 The Authors. APMIS published by John Wiley & Sons Ltd on behalf of Scandinavian Societies for Medical Microbiology and Pathology.
Figures



Similar articles
-
Assessment of Treatment-Relevant Immune Biomarkers in Psoriasis and Atopic Dermatitis: Toward Personalized Medicine in Dermatology.J Invest Dermatol. 2023 Aug;143(8):1412-1422. doi: 10.1016/j.jid.2023.04.005. Epub 2023 Jun 20. J Invest Dermatol. 2023. PMID: 37341663 Free PMC article. Review.
-
Multi-Omics Approach to Improved Diagnosis and Treatment of Atopic Dermatitis and Psoriasis.Int J Mol Sci. 2024 Jan 15;25(2):1042. doi: 10.3390/ijms25021042. Int J Mol Sci. 2024. PMID: 38256115 Free PMC article. Review.
-
Toward Precision Medicine in Atopic Dermatitis Using Molecular-Based Approaches.Actas Dermosifiliogr. 2024 Jan;115(1):66-75. doi: 10.1016/j.ad.2023.08.003. Epub 2023 Aug 29. Actas Dermosifiliogr. 2024. PMID: 37652096 Review. English, Spanish.
-
Atopic dermatitis endotypes: knowledge for personalized medicine.Curr Opin Allergy Clin Immunol. 2022 Jun 1;22(3):153-159. doi: 10.1097/ACI.0000000000000820. Epub 2022 Feb 11. Curr Opin Allergy Clin Immunol. 2022. PMID: 35152229 Review.
-
Atopic dermatitis stratification: current and future perspective on skin and blood transcriptomic and proteomic profiling.Expert Rev Clin Immunol. 2024 Sep;20(9):1083-1088. doi: 10.1080/1744666X.2024.2323964. Epub 2024 Mar 4. Expert Rev Clin Immunol. 2024. PMID: 38436065 Review.
Cited by
-
The Translational Dermatology Initiative: Aiming at a New Disease Classification of Inflammatory Skin Diseases.JID Innov. 2025 May 13;5(5):100381. doi: 10.1016/j.xjidi.2025.100381. eCollection 2025 Sep. JID Innov. 2025. PMID: 40535547 Free PMC article.
-
Next-Generation Oral Delivery Systems: Phytosomal Hinokitiol Tablets via REGEMAT 3D Bioprinter-Based 3D Printing for Enhanced Bioavailability.Scientifica (Cairo). 2025 Jun 30;2025:6678786. doi: 10.1155/sci5/6678786. eCollection 2025. Scientifica (Cairo). 2025. PMID: 40661180 Free PMC article.
-
Review of Personalized Medicine and Pharmacogenomics of Anti-Cancer Compounds and Natural Products.Genes (Basel). 2024 Apr 8;15(4):468. doi: 10.3390/genes15040468. Genes (Basel). 2024. PMID: 38674402 Free PMC article. Review.
-
Overview of omics biomarkers in pituitary neuroendocrine tumors to design future diagnosis and treatment strategies.EPMA J. 2021 Jun 26;12(3):383-401. doi: 10.1007/s13167-021-00246-1. eCollection 2021 Sep. EPMA J. 2021. PMID: 34567287 Free PMC article. Review.
-
Omics-Driven Biomarkers of Psoriasis: Recent Insights, Current Challenges, and Future Prospects.Clin Cosmet Investig Dermatol. 2020 Aug 25;13:611-625. doi: 10.2147/CCID.S227896. eCollection 2020. Clin Cosmet Investig Dermatol. 2020. PMID: 32922059 Free PMC article. Review.
References
-
- Vail J. Pharmacogenomics: the end of trial‐and‐error medicine? Int J Pharm Compd 2007;11:59–65. - PubMed
-
- Morgan P, Brown DG, Lennard S, Anderton MJ, Barrett JC, Eriksson U, et al. Impact of a five‐dimensional framework on R&D productivity at AstraZeneca. Nat Rev Drug Discov 2018;17:167–81. - PubMed
-
- Rhen T, Cidlowski JA. Antiinflammatory action of glucocorticoids–new mechanisms for old drugs. N Engl J Med 2005;353:1711–23. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

