Live-cell imaging reveals enhancer-dependent Sox2 transcription in the absence of enhancer proximity
- PMID: 31124784
- PMCID: PMC6534382
- DOI: 10.7554/eLife.41769
Live-cell imaging reveals enhancer-dependent Sox2 transcription in the absence of enhancer proximity
Abstract
Enhancers are important regulatory elements that can control gene activity across vast genetic distances. However, the underlying nature of this regulation remains obscured because it has been difficult to observe in living cells. Here, we visualize the spatial organization and transcriptional output of the key pluripotency regulator Sox2 and its essential enhancer Sox2 Control Region (SCR) in living embryonic stem cells (ESCs). We find that Sox2 and SCR show no evidence of enhanced spatial proximity and that spatial dynamics of this pair is limited over tens of minutes. Sox2 transcription occurs in short, intermittent bursts in ESCs and, intriguingly, we find this activity demonstrates no association with enhancer proximity, suggesting that direct enhancer-promoter contacts do not drive contemporaneous Sox2 transcription. Our study establishes a framework for interrogation of enhancer function in living cells and supports an unexpected mechanism for enhancer control of Sox2 expression that uncouples transcription from enhancer proximity.
Keywords: chromosomes; enhancers; gene expression; microscopy; mouse; transcription.
© 2019, Alexander et al.
Conflict of interest statement
JA, JG, BL, LM, MS, YS, BH, SL, OW No competing interests declared
Figures
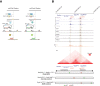

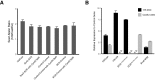
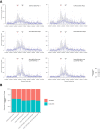
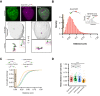





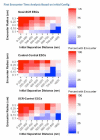

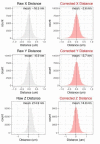

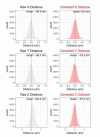
References
-
- Afgan E, Baker D, Batut B, van den Beek M, Bouvier D, Cech M, Chilton J, Clements D, Coraor N, Grüning BA, Guerler A, Hillman-Jackson J, Hiltemann S, Jalili V, Rasche H, Soranzo N, Goecks J, Taylor J, Nekrutenko A, Blankenberg D. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Research. 2018;46:W537–W544. doi: 10.1093/nar/gky379. - DOI - PMC - PubMed
-
- Alberts B, Johnson AD, Lewis J, Morgan D, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. W. W. Norton & Company; 2014.
-
- Alexander JM. Alexander_et_al_2018. f8a68bcGitHub. 2018 https://github.com/JMAlexander/2018_eLife_Alexander_et_al
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
- Medical Research Grant/W. M. Keck Foundation/International
- R21 EB021453/EB/NIBIB NIH HHS/United States
- 16POST309100006/American Heart Association/International
- R35GM118167/GM/NIGMS NIH HHS/United States
- R01 DC013560/DC/NIDCD NIH HHS/United States
- UM1 HG009402/HG/NHGRI NIH HHS/United States
- R01DC013560/DC/NIDCD NIH HHS/United States
- T32GM007175/GM/NIGMS NIH HHS/United States
- 16POST30910006/AHA/American Heart Association-American Stroke Association/United States
- R21HG010065/HG/NHGRI NIH HHS/United States
- UM1HG009402/HG/NHGRI NIH HHS/United States
- R21 HG010065/HG/NHGRI NIH HHS/United States
- R21EB021453/EB/NIBIB NIH HHS/United States
- R21EB022787/EB/NIBIB NIH HHS/United States
- R21 EB022787/EB/NIBIB NIH HHS/United States
- R35 GM118167/GM/NIGMS NIH HHS/United States
- T32 GM007175/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

