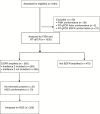
Defining EGFR amplification status for clinical trial inclusion
- PMID: 31125418
- PMCID: PMC6784284
- DOI: 10.1093/neuonc/noz096
Defining EGFR amplification status for clinical trial inclusion
Abstract
Background: Precision medicine trials targeting the epidermal growth factor receptor (EGFR) in glioblastoma patients require selection for EGFR-amplified tumors. However, there is currently no gold standard in determining the amplification status of EGFR or variant III (EGFRvIII) expression. Here, we aimed to determine which technique and which cutoffs are suitable to determine EGFR amplification status.
Methods: We compared fluorescence in-situ hybridization (FISH) and real-time quantitative (RT-q)PCR data from patients screened for trial inclusion into the Intellance 2 clinical trial, with data from a panel-based next generation sequencing (NGS) platform (both DNA and RNA).
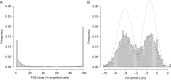
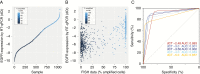
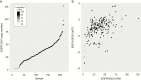
Results: By using data from >1000 samples, we show that at least 50% of EGFR amplified nuclei should be present to define EGFR gene amplification by FISH. Gene amplification (as determined by FISH) correlates with EGFR expression levels (as determined by RT-qPCR) with receiver operating characteristics analysis showing an area under the curve of up to 0.902. EGFR expression as assessed by RT-qPCR therefore may function as a surrogate marker for EGFR amplification. Our NGS data show that EGFR copy numbers can strongly vary between tumors, with levels ranging from 2 to more than 100 copies per cell. Levels exceeding 5 gene copies can be used to define EGFR-amplification by NGS; below this level, FISH detects very few (if any) EGFR amplified nuclei and none of the samples express EGFRvIII.
Conclusion: Our data from central laboratories and diagnostic sequencing facilities, using material from patients eligible for clinical trial inclusion, help define the optimal cutoff for various techniques to determine EGFR amplification for diagnostic purposes.
Keywords: EGFR; EGFRvIII; FISH; amplification; biomarker; glioblastoma; screening.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Society for Neuro-Oncology.
Figures


Comment in
-
EGFR amplification status for clinical trial inclusion: where do we draw the line?Neuro Oncol. 2019 Oct 9;21(10):1215-1216. doi: 10.1093/neuonc/noz146. Neuro Oncol. 2019. PMID: 31504815 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

