Dysfunction in the arbuscular mycorrhizal symbiosis has consistent but small effects on the establishment of the fungal microbiota in Lotus japonicus
- PMID: 31125425
- PMCID: PMC6773208
- DOI: 10.1111/nph.15958
Dysfunction in the arbuscular mycorrhizal symbiosis has consistent but small effects on the establishment of the fungal microbiota in Lotus japonicus
Abstract
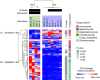
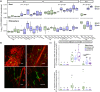
Most land plants establish mutualistic interactions with arbuscular mycorrhizal (AM) fungi. Intracellular accommodation of AM fungal symbionts remodels important host traits like root morphology and nutrient acquisition. How mycorrhizal colonization impacts plant microbiota is unclear. To understand the impact of AM symbiosis on fungal microbiota, ten Lotus japonicus mutants impaired at different stages of AM formation were grown in non-sterile natural soil and their root-associated fungal communities were studied. Plant mutants lacking the capacity to form mature arbuscules (arb- ) exhibited limited growth performance associated with altered phosphorus (P) acquisition and reduction-oxidation (redox) processes. Furthermore, arb- plants assembled moderately but consistently different root-associated fungal microbiota, characterized by the depletion of Glomeromycota and the concomitant enrichment of Ascomycota, including Dactylonectria torresensis. Single and co-inoculation experiments showed a strong reduction of root colonization by D. torresensis in the presence of AM fungus Rhizophagus irregularis, particularly in arbuscule-forming plants. Our results suggest that impairment of central symbiotic functions in AM host plants leads to specific changes in root microbiomes and in tripartite interactions between the host plant, AM and non-AM fungi. This lays the foundation for mechanistic studies on microbe-microbe and microbe-host interactions in AM symbiosis of the model L. japonicus.
Keywords: RNA-seq; arbuscular mycorrhizal (AM) fungi; fungal community; legume; natural soil; symbiosis.
© 2019 The Authors. New Phytologist © 2019 New Phytologist Trust.
Figures



Similar articles
-
Lotus japonicus Symbiosis Genes Impact Microbial Interactions between Symbionts and Multikingdom Commensal Communities.mBio. 2019 Oct 8;10(5):e01833-19. doi: 10.1128/mBio.01833-19. mBio. 2019. PMID: 31594815 Free PMC article.
-
Gibberellins interfere with symbiosis signaling and gene expression and alter colonization by arbuscular mycorrhizal fungi in Lotus japonicus.Plant Physiol. 2015 Feb;167(2):545-57. doi: 10.1104/pp.114.247700. Epub 2014 Dec 19. Plant Physiol. 2015. PMID: 25527715 Free PMC article.
-
Isolation and phenotypic characterization of Lotus japonicus mutants specifically defective in arbuscular mycorrhizal formation.Plant Cell Physiol. 2014 May;55(5):928-41. doi: 10.1093/pcp/pcu024. Epub 2014 Feb 2. Plant Cell Physiol. 2014. PMID: 24492255
-
A journey into the world of small RNAs in the arbuscular mycorrhizal symbiosis.New Phytol. 2024 May;242(4):1534-1544. doi: 10.1111/nph.19394. Epub 2023 Nov 20. New Phytol. 2024. PMID: 37985403 Review.
-
The arbuscular mycorrhizal symbiosis: a molecular review of the fungal dimension.J Exp Bot. 2001 Mar;52(Spec Issue):469-78. doi: 10.1093/jexbot/52.suppl_1.469. J Exp Bot. 2001. PMID: 11326053 Review.
Cited by
-
Rhizosphere frame system enables nondestructive live-imaging of legume-rhizobium interactions in the soil.J Plant Res. 2023 Sep;136(5):769-780. doi: 10.1007/s10265-023-01476-2. Epub 2023 Jul 4. J Plant Res. 2023. PMID: 37402088 Free PMC article.
-
Receptor-Like Kinases Sustain Symbiotic Scrutiny.Plant Physiol. 2020 Apr;182(4):1597-1612. doi: 10.1104/pp.19.01341. Epub 2020 Feb 13. Plant Physiol. 2020. PMID: 32054781 Free PMC article. Review.
-
Lotus japonicus Symbiosis Genes Impact Microbial Interactions between Symbionts and Multikingdom Commensal Communities.mBio. 2019 Oct 8;10(5):e01833-19. doi: 10.1128/mBio.01833-19. mBio. 2019. PMID: 31594815 Free PMC article.
-
The genetic architecture of host response reveals the importance of arbuscular mycorrhizae to maize cultivation.Elife. 2020 Nov 19;9:e61701. doi: 10.7554/eLife.61701. Elife. 2020. PMID: 33211006 Free PMC article.
-
Neighboring plants divergently modulate effects of loss-of-function in maize mycorrhizal phosphate uptake on host physiology and root fungal microbiota.PLoS One. 2020 Jun 17;15(6):e0232633. doi: 10.1371/journal.pone.0232633. eCollection 2020. PLoS One. 2020. PMID: 32555651 Free PMC article.
References
-
- Agustí‐Brisach C, Mostert L, Armengol J. 2014. Detection and quantification of Ilyonectria spp. associated with black‐foot disease of grapevine in nursery soils using multiplex nested PCR and quantitative PCR. Plant Pathology 63: 316–322.
-
- Azcón‐Aguilar C, Barea J. 1997. Arbuscular mycorrhizas and biological control of soil‐borne plant pathogens – an overview of the mechanisms involved. Mycorrhiza 6: 457–464.
-
- Bansal M, Mukerji K. 1994. Positive correlation between VAM‐induced changes in root exudation and mycorrhizosphere mycoflora. Mycorrhiza 5: 39–44.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

