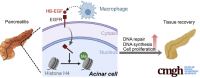
Myeloid Cell-Derived HB-EGF Drives Tissue Recovery After Pancreatitis
- PMID: 31125624
- PMCID: PMC6661420
- DOI: 10.1016/j.jcmgh.2019.05.006
Myeloid Cell-Derived HB-EGF Drives Tissue Recovery After Pancreatitis
Abstract
Background & aims: Pancreatitis is a major cause of morbidity and mortality and is a risk factor for pancreatic tumorigenesis. Upon tissue damage, an inflammatory response, made up largely of macrophages, provides multiple growth factors that promote repair. Here, we examine the molecular pathways initiated by macrophages to promote pancreas recovery from pancreatitis.
Methods: To induce organ damage, mice were subjected to cerulein-induced experimental pancreatitis and analyzed at various times of recovery. CD11b-DTR mice were used to deplete myeloid cells. Hbegff/f;LysM-Cre mice were used to ablate myeloid cell-derived heparin-binding epidermal growth factor (EGF)-like growth factor (HB-EGF). To ablate EGFR specifically during recovery, pancreatitis was induced in Egfrf/f;Ptf1aFlpO/+;FSF-Rosa26CAG-CreERT2 mice followed by tamoxifen treatment.
Results: Macrophages infiltrating the pancreas in experimental pancreatitis make high levels of HB-EGF. Both depletion of myeloid cells and ablation of myeloid cell HB-EGF delayed recovery from experimental pancreatitis, resulting from a decrease in cell proliferation and an increase in apoptosis. Mechanistically, ablation of myeloid cell HB-EGF impaired epithelial cell DNA repair, ultimately leading to cell death. Soluble HB-EGF induced EGFR nuclear translocation and methylation of histone H4, facilitating resolution of DNA damage in pancreatic acinar cells in vitro. Consistent with its role as the primary receptor of HB-EGF, in vivo ablation of EGFR from pancreatic epithelium during recovery from pancreatitis resulted in accumulation of DNA damage.
Conclusions: By using novel conditional knockout mouse models, we determined that HB-EGF derived exclusively from myeloid cells induces epithelial cell proliferation and EGFR-dependent DNA repair, facilitating pancreas healing after injury.
Keywords: DNA Damage; EGFR; Inflammation; Macrophages.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Figures






Similar articles
-
Myeloid- and Epithelial-derived Heparin-Binding Epidermal Growth Factor-like Growth Factor Promotes Pulmonary Fibrosis.Am J Respir Cell Mol Biol. 2022 Dec;67(6):641-653. doi: 10.1165/rcmb.2022-0174OC. Am J Respir Cell Mol Biol. 2022. PMID: 36036796 Free PMC article.
-
Activation of Epidermal Growth Factor Receptor in Macrophages Mediates Feedback Inhibition of M2 Polarization and Gastrointestinal Tumor Cell Growth.J Biol Chem. 2016 Sep 23;291(39):20462-72. doi: 10.1074/jbc.M116.750182. Epub 2016 Aug 9. J Biol Chem. 2016. PMID: 27507810 Free PMC article.
-
Arsenic Attenuates Heparin-Binding EGF-Like Growth Factor/EGFR Signaling That Promotes Matrix Metalloprotease 9-Dependent Astrocyte Damage in the Developing Rat Brain.Toxicol Sci. 2018 Apr 1;162(2):406-428. doi: 10.1093/toxsci/kfx264. Toxicol Sci. 2018. PMID: 29228391
-
Heparin-Binding Epidermal Growth Factor-Like Growth Factor as a Critical Mediator of Tissue Repair and Regeneration.Am J Pathol. 2018 Nov;188(11):2446-2456. doi: 10.1016/j.ajpath.2018.07.016. Epub 2018 Aug 22. Am J Pathol. 2018. PMID: 30142332 Free PMC article. Review.
-
Heparin-binding epidermal growth factor-like growth factor (HB-EGF) and proteolytic processing by a disintegrin and metalloproteinases (ADAM): a regulator of several pathways.Semin Cell Dev Biol. 2014 Apr;28:22-30. doi: 10.1016/j.semcdb.2014.03.004. Epub 2014 Mar 25. Semin Cell Dev Biol. 2014. PMID: 24680771 Review.
Cited by
-
Arginase 1 is a key driver of immune suppression in pancreatic cancer.Elife. 2023 Feb 2;12:e80721. doi: 10.7554/eLife.80721. Elife. 2023. PMID: 36727849 Free PMC article.
-
Pancreatic Fibroblast Heterogeneity: From Development to Cancer.Cells. 2020 Nov 12;9(11):2464. doi: 10.3390/cells9112464. Cells. 2020. PMID: 33198201 Free PMC article. Review.
-
Tuft cells transdifferentiate to neural-like progenitor cells in the progression of pancreatic cancer.bioRxiv [Preprint]. 2024 Apr 23:2024.02.12.579982. doi: 10.1101/2024.02.12.579982. bioRxiv. 2024. Update in: Dev Cell. 2025 Mar 24;60(6):837-852.e3. doi: 10.1016/j.devcel.2024.12.003. PMID: 38405804 Free PMC article. Updated. Preprint.
-
Tuft cells transdifferentiate to neural-like progenitor cells in the progression of pancreatic cancer.Dev Cell. 2025 Mar 24;60(6):837-852.e3. doi: 10.1016/j.devcel.2024.12.003. Epub 2024 Dec 24. Dev Cell. 2025. PMID: 39721583
-
Novel IL-4/HB-EGF-dependent crosstalk between eosinophils and macrophages controls liver regeneration after ischaemia and reperfusion injury.Gut. 2024 Aug 8;73(9):1543-1553. doi: 10.1136/gutjnl-2024-332033. Gut. 2024. PMID: 38724220 Free PMC article.
References
-
- Kleeff J., Whitcomb D.C., Shimosegawa T., Esposito I., Lerch M.M., Gress T., Mayerle J., Drewes A.M., Rebours V., Akisik F., Munoz J.E.D., Neoptolemos J.P. Chronic pancreatitis. Nat Rev Dis Primers. 2017;3:17060. - PubMed
-
- Seifert L., Werba G., Tiwari S., Giao Ly N.N., Alothman S., Alqunaibit D., Avanzi A., Barilla R., Daley D., Greco S.H., Torres-Hernandez A., Pergamo M., Ochi A., Zambirinis C.P., Pansari M., Rendon M., Tippens D., Hundeyin M., Mani V.R., Hajdu C., Engle D., Miller G. The necrosome promotes pancreatic oncogenesis via CXCL1 and Mincle-induced immune suppression. Nature. 2016;532:245–249. - PMC - PubMed
-
- Bockman D.E., Boydston W.R., Anderson M.C. Origin of tubular complexes in human chronic pancreatitis. Am J Surg. 1982;144:243–249. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 CA198074/CA/NCI NIH HHS/United States
- P30 CA046592/CA/NCI NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- U01 DK089570/DK/NIDDK NIH HHS/United States
- U2C DK059637/DK/NIDDK NIH HHS/United States
- U01 DK072473/DK/NIDDK NIH HHS/United States
- R01 CA151588/CA/NCI NIH HHS/United States
- R01 CA159222/CA/NCI NIH HHS/United States
- P30 DK020593/DK/NIDDK NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- K08 CA201581/CA/NCI NIH HHS/United States
- U01 CA224145/CA/NCI NIH HHS/United States
- U24 DK059637/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

