Effects of KEAP1 Silencing on the Regulation of NRF2 Activity in Neuroendocrine Lung Tumors
- PMID: 31126053
- PMCID: PMC6566555
- DOI: 10.3390/ijms20102531
Effects of KEAP1 Silencing on the Regulation of NRF2 Activity in Neuroendocrine Lung Tumors
Abstract
Background: The KEAP1/NRF2 pathway has been widely investigated in tumors since it was implicated in cancer cells survival and therapies resistance. In lung tumors the deregulation of this pathway is mainly related to point mutations of KEAP1 and NFE2L2 genes and KEAP1 promoter hypermethylation, but these two genes have been rarely investigated in low/intermediate grade neuroendocrine tumors of the lung.
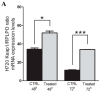
Methods: The effects of KEAP1 silencing on NRF2 activity was investigated in H720 and H727 carcinoid cell lines and results were compared with those obtained by molecular profiling of KEAP1 and NFE2L2 in a collection of 47 lung carcinoids. The correlation between methylation and transcript levels was assessed by 5-aza-dC treatment.
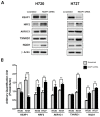
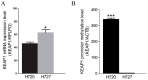
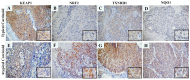
Results: We demonstrated that in carcinoid cell lines, the KEAP1 silencing induces an upregulation of NRF2 and some of its targets and that there is a direct correlation between KEAP1 methylation and its mRNA levels. A KEAP1 hypermethylation and Loss of Heterozygosity at KEAP1 gene locus was also observed in nearly half of lung carcinoids.
Conclusions: This is the first study that has described the effects of KEAP1 silencing on the regulation of NRF2 activity in lung carcinoids cells. The epigenetic deregulation of the KEAP1/NRF2 by a KEAP1 promoter hypermethylation system appears to be a frequent event in lung carcinoids.
Keywords: KEAP1; Lung Carcinoid; NRF2; methylation; mutation; outcome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Keap1/Nrf2 pathway in kidney cancer: frequent methylation of KEAP1 gene promoter in clear renal cell carcinoma.Oncotarget. 2017 Feb 14;8(7):11187-11198. doi: 10.18632/oncotarget.14492. Oncotarget. 2017. PMID: 28061437 Free PMC article.
-
Methylation Density Pattern of KEAP1 Gene in Lung Cancer Cell Lines Detected by Quantitative Methylation Specific PCR and Pyrosequencing.Int J Mol Sci. 2019 May 31;20(11):2697. doi: 10.3390/ijms20112697. Int J Mol Sci. 2019. PMID: 31159323 Free PMC article.
-
Clinicopathological, microenvironmental and genetic determinants of molecular subtypes in KEAP1/NRF2-mutant lung cancer.Int J Cancer. 2019 Feb 15;144(4):788-801. doi: 10.1002/ijc.31975. Epub 2018 Dec 4. Int J Cancer. 2019. PMID: 30411339
-
Epigenetic versus Genetic Deregulation of the KEAP1/NRF2 Axis in Solid Tumors: Focus on Methylation and Noncoding RNAs.Oxid Med Cell Longev. 2018 Mar 13;2018:2492063. doi: 10.1155/2018/2492063. eCollection 2018. Oxid Med Cell Longev. 2018. PMID: 29643973 Free PMC article. Review.
-
Genetic and epigenetic regulation of the NRF2-KEAP1 pathway in human lung cancer.Br J Cancer. 2022 May;126(9):1244-1252. doi: 10.1038/s41416-021-01642-0. Epub 2021 Nov 29. Br J Cancer. 2022. PMID: 34845361 Free PMC article. Review.
Cited by
-
Identifying General Tumor and Specific Lung Cancer Biomarkers by Transcriptomic Analysis.Biology (Basel). 2022 Jul 20;11(7):1082. doi: 10.3390/biology11071082. Biology (Basel). 2022. PMID: 36101460 Free PMC article.
-
Lung neuroendocrine tumors: A systematic literature review (Review).Exp Ther Med. 2022 Feb;23(2):176. doi: 10.3892/etm.2021.11099. Epub 2021 Dec 28. Exp Ther Med. 2022. PMID: 35069857 Free PMC article. Review.
-
Role of NRF2 in Lung Cancer.Cells. 2021 Jul 24;10(8):1879. doi: 10.3390/cells10081879. Cells. 2021. PMID: 34440648 Free PMC article. Review.
-
CRISPR-Generated Nrf2a Loss- and Gain-of-Function Mutants Facilitate Mechanistic Analysis of Chemical Oxidative Stress-Mediated Toxicity in Zebrafish.Chem Res Toxicol. 2020 Feb 17;33(2):426-435. doi: 10.1021/acs.chemrestox.9b00346. Epub 2020 Jan 8. Chem Res Toxicol. 2020. PMID: 31858786 Free PMC article.
-
Molecular Pathology of Well-Differentiated Pulmonary and Thymic Neuroendocrine Tumors: What Do Pathologists Need to Know?Endocr Pathol. 2021 Mar;32(1):154-168. doi: 10.1007/s12022-021-09668-z. Epub 2021 Feb 27. Endocr Pathol. 2021. PMID: 33641055 Free PMC article. Review.
References
-
- Frank R., Scheffler M., Merkelbach-Bruse S., Ihle M.A., Kron A., Rauer M., Ueckeroth F., König K., Michels S., Fischer R., et al. Clinical and Pathological Characteristics of KEAP1- and NFE2L2-Mutated Non-Small Cell Lung Carcinoma (NSCLC) Clin. Cancer Res. 2018;24:3087–3096. doi: 10.1158/1078-0432.CCR-17-3416. - DOI - PubMed

