Bif-1 Interacts with Prohibitin-2 to Regulate Mitochondrial Inner Membrane during Cell Stress and Apoptosis
- PMID: 31126972
- PMCID: PMC6622411
- DOI: 10.1681/ASN.2018111117
Bif-1 Interacts with Prohibitin-2 to Regulate Mitochondrial Inner Membrane during Cell Stress and Apoptosis
Abstract
Background: Mitochondria are dynamic organelles that undergo fission and fusion. During cell stress, mitochondrial dynamics shift to fission, leading to mitochondrial fragmentation, membrane leakage, and apoptosis. Mitochondrial fragmentation requires the cleavage of both outer and inner membranes, but the mechanism of inner membrane cleavage is unclear. Bif-1 and prohibitin-2 may regulate mitochondrial dynamics.
Methods: We used azide-induced ATP depletion to incite cell stress in mouse embryonic fibroblasts and renal proximal tubular cells, and renal ischemia-reperfusion to induce stress in mice. We also used knockout cells and mice to determine the role of Bif-1, and used multiple techniques to analyze the molecular interaction between Bif-1 and prohibitin-2.
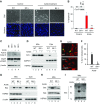
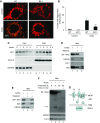
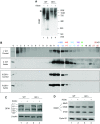
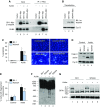
Results: Upon cell stress, Bif-1 translocated to mitochondria to bind prohibitin-2, resulting in the disruption of prohibitin complex and proteolytic inactivation of the inner membrane fusion protein OPA1. Bif-1-deficiency inhibited prohibitin complex disruption, OPA1 proteolysis, mitochondrial fragmentation, and apoptosis. Domain deletion analysis indicated that Bif-1 interacted with prohibitin-2 via its C-terminus. Notably, mutation of Bif-1 at its C-terminal tryptophan-344 not only prevented Bif-1/prohibitin-2 interaction but also reduced prohibitin complex disruption, OPA1 proteolysis, mitochondrial fragmentation, and apoptosis, supporting a pathogenic role of Bif-1/prohibitin-2 interaction. In mice, Bif-1 bound prohibitin-2 during renal ischemia/reperfusion injury, and Bif-1-deficiency protected against OPA1 proteolysis, mitochondrial fragmentation, apoptosis and kidney injury.
Conclusions: These findings suggest that during cell stress, Bif-1 regulates mitochondrial inner membrane by interacting with prohibitin-2 to disrupt prohibitin complexes and induce OPA1 proteolysis and inactivation.
Keywords: apoptosis; mitochondria; renal ischemia.
Copyright © 2019 by the American Society of Nephrology.
Figures

Similar articles
-
OMA1 mediates OPA1 proteolysis and mitochondrial fragmentation in experimental models of ischemic kidney injury.Am J Physiol Renal Physiol. 2014 Jun 1;306(11):F1318-26. doi: 10.1152/ajprenal.00036.2014. Epub 2014 Mar 26. Am J Physiol Renal Physiol. 2014. PMID: 24671334 Free PMC article.
-
Membrane depolarization activates the mitochondrial protease OMA1 by stimulating self-cleavage.EMBO Rep. 2014 May;15(5):576-85. doi: 10.1002/embr.201338240. Epub 2014 Apr 9. EMBO Rep. 2014. PMID: 24719224 Free PMC article.
-
In vivo stabilization of OPA1 in hepatocytes potentiates mitochondrial respiration and gluconeogenesis in a prohibitin-dependent way.J Biol Chem. 2019 Aug 23;294(34):12581-12598. doi: 10.1074/jbc.RA119.007601. Epub 2019 Jul 8. J Biol Chem. 2019. PMID: 31285263 Free PMC article.
-
OPA1 processing in cell death and disease - the long and short of it.J Cell Sci. 2016 Jun 15;129(12):2297-306. doi: 10.1242/jcs.159186. Epub 2016 May 17. J Cell Sci. 2016. PMID: 27189080 Review.
-
Targeted OMA1 therapies for cancer.Int J Cancer. 2019 Nov 1;145(9):2330-2341. doi: 10.1002/ijc.32177. Epub 2019 Feb 21. Int J Cancer. 2019. PMID: 30714136 Review.
Cited by
-
Mitochondrial quality control in human health and disease.Mil Med Res. 2024 May 29;11(1):32. doi: 10.1186/s40779-024-00536-5. Mil Med Res. 2024. PMID: 38812059 Free PMC article. Review.
-
The function of prohibitins in mitochondria and the clinical potentials.Cancer Cell Int. 2022 Nov 8;22(1):343. doi: 10.1186/s12935-022-02765-x. Cancer Cell Int. 2022. PMID: 36348375 Free PMC article. Review.
-
Bax inhibitor 1 preserves mitochondrial homeostasis in acute kidney injury through promoting mitochondrial retention of PHB2.Theranostics. 2020 Jan 1;10(1):384-397. doi: 10.7150/thno.40098. eCollection 2020. Theranostics. 2020. PMID: 31903127 Free PMC article.
-
Unveiling the potential of mitochondrial dynamics as a therapeutic strategy for acute kidney injury.Front Cell Dev Biol. 2023 Aug 11;11:1244313. doi: 10.3389/fcell.2023.1244313. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 37635869 Free PMC article. Review.
-
The Ethyl Acetate Extract From Celastrus orbiculatus Promotes Apoptosis of Gastric Cancer Cells Through Mitochondria Regulation by PHB.Front Pharmacol. 2021 May 28;12:635467. doi: 10.3389/fphar.2021.635467. eCollection 2021. Front Pharmacol. 2021. PMID: 34122065 Free PMC article.
References
-
- Chan DC: Fusion and fission: Interlinked Processes critical for mitochondrial health. Annu Rev Genet 46: 265–287, 2012 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

