Mosaic origin of the eukaryotic kinetochore
- PMID: 31127038
- PMCID: PMC6601020
- DOI: 10.1073/pnas.1821945116
Mosaic origin of the eukaryotic kinetochore
Abstract
The emergence of eukaryotes from ancient prokaryotic lineages embodied a remarkable increase in cellular complexity. While prokaryotes operate simple systems to connect DNA to the segregation machinery during cell division, eukaryotes use a highly complex protein assembly known as the kinetochore. Although conceptually similar, prokaryotic segregation systems and the eukaryotic kinetochore are not homologous. Here we investigate the origins of the kinetochore before the last eukaryotic common ancestor (LECA) using phylogenetic trees, sensitive profile-versus-profile homology detection, and structural comparisons of its protein components. We show that LECA's kinetochore proteins share deep evolutionary histories with proteins involved in a few prokaryotic systems and a multitude of eukaryotic processes, including ubiquitination, transcription, and flagellar and vesicular transport systems. We find that gene duplications played a major role in shaping the kinetochore; more than half of LECA's kinetochore proteins have other kinetochore proteins as closest homologs. Some of these have no detectable homology to any other eukaryotic protein, suggesting that they arose as kinetochore-specific folds before LECA. We propose that the primordial kinetochore evolved from proteins involved in various (pre)eukaryotic systems as well as evolutionarily novel folds, after which a subset duplicated to give rise to the complex kinetochore of LECA.
Keywords: LECA; eukaryogenesis; gene duplication; kinetochore; mitosis.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



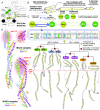
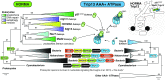
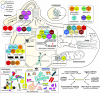
Comment in
-
The kinetochore and the origin of eukaryotic chromosome segregation.Proc Natl Acad Sci U S A. 2019 Jun 25;116(26):12596-12598. doi: 10.1073/pnas.1908067116. Epub 2019 Jun 7. Proc Natl Acad Sci U S A. 2019. PMID: 31175149 Free PMC article. No abstract available.
References
-
- Henikoff S., Ahmad K., Malik H. S., The centromere paradox: Stable inheritance with rapidly evolving DNA. Science 293, 1098–1102 (2001). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

