Activation of Peroxisome Proliferator-Activated Receptor-α Increases the Expression of Nuclear Receptor Related 1 Protein (Nurr1) in Dopaminergic Neurons
- PMID: 31127527
- PMCID: PMC8174404
- DOI: 10.1007/s12035-019-01649-y
Activation of Peroxisome Proliferator-Activated Receptor-α Increases the Expression of Nuclear Receptor Related 1 Protein (Nurr1) in Dopaminergic Neurons
Abstract
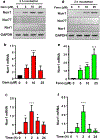
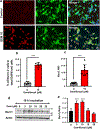
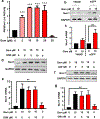
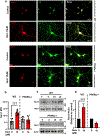
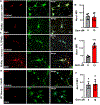
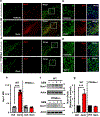
Nuclear receptor related 1 protein (Nurr1) is an important transcription factor required for differentiation and maintenance of midbrain dopaminergic (DA) neurons. Since decrease in Nurr1 function either due to diminished expression or rare mutation is associated with Parkinson's disease (PD), upregulation of Nurr1 may be beneficial for PD. However, such mechanisms are poorly understood. This study underlines the importance of peroxisome proliferator-activated receptor (PPAR)α in controlling the transcription of Nurr1. Our mRNA analyses followed by different immunoassays clearly indicated that PPARα agonist gemfibrozil strongly upregulated the expression of Nurr1 in wild-type, but not PPARα-/-, DA neurons. Moreover, identification of conserved PPRE in the promoter of Nurr1 gene followed by chromatin immunoprecipitation analysis, PPRE luciferase assay, and manipulation of Nurr1 gene by viral transduction of different PPARα plasmids confirmed that PPARα was indeed involved in the expression of Nurr1. Finally, oral administration of gemfibrozil increased Nurr1 expression in vivo in nigra of wild-type, but not PPARα-/-, mice identifying PPARα as a novel regulator of Nurr1 expression and associated protection of DA neurons.
Keywords: Dopaminergic neurons; Gemfibrozil; Nurr1; PPARα; Parkinson’s disease.
Figures
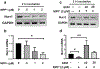

Similar articles
-
Gemfibrozil Protects Dopaminergic Neurons in a Mouse Model of Parkinson's Disease via PPARα-Dependent Astrocytic GDNF Pathway.J Neurosci. 2021 Mar 10;41(10):2287-2300. doi: 10.1523/JNEUROSCI.3018-19.2021. Epub 2021 Jan 29. J Neurosci. 2021. PMID: 33514677 Free PMC article.
-
Gemfibrozil and fenofibrate, Food and Drug Administration-approved lipid-lowering drugs, up-regulate tripeptidyl-peptidase 1 in brain cells via peroxisome proliferator-activated receptor α: implications for late infantile Batten disease therapy.J Biol Chem. 2012 Nov 9;287(46):38922-35. doi: 10.1074/jbc.M112.365148. Epub 2012 Sep 18. J Biol Chem. 2012. PMID: 22989886 Free PMC article.
-
Lethal Factor Domain-Mediated Delivery of Nurr1 Transcription Factor Enhances Tyrosine Hydroxylase Activity and Protects from Neurotoxin-Induced Degeneration of Dopaminergic Cells.Mol Neurobiol. 2019 May;56(5):3393-3403. doi: 10.1007/s12035-018-1311-6. Epub 2018 Aug 18. Mol Neurobiol. 2019. PMID: 30121937 Free PMC article.
-
NURR1 in Parkinson disease--from pathogenesis to therapeutic potential.Nat Rev Neurol. 2013 Nov;9(11):629-36. doi: 10.1038/nrneurol.2013.209. Epub 2013 Oct 15. Nat Rev Neurol. 2013. PMID: 24126627 Review.
-
The lifelong maintenance of mesencephalic dopaminergic neurons by Nurr1 and engrailed.J Biomed Sci. 2014 Apr 1;21(1):27. doi: 10.1186/1423-0127-21-27. J Biomed Sci. 2014. PMID: 24685177 Free PMC article. Review.
Cited by
-
Selective targeting of the TLR2/MyD88/NF-κB pathway reduces α-synuclein spreading in vitro and in vivo.Nat Commun. 2021 Sep 10;12(1):5382. doi: 10.1038/s41467-021-25767-1. Nat Commun. 2021. PMID: 34508096 Free PMC article.
-
Gemfibrozil Protects Dopaminergic Neurons in a Mouse Model of Parkinson's Disease via PPARα-Dependent Astrocytic GDNF Pathway.J Neurosci. 2021 Mar 10;41(10):2287-2300. doi: 10.1523/JNEUROSCI.3018-19.2021. Epub 2021 Jan 29. J Neurosci. 2021. PMID: 33514677 Free PMC article.
-
Selective Inhibition of the Interaction between SARS-CoV-2 Spike S1 and ACE2 by SPIDAR Peptide Induces Anti-Inflammatory Therapeutic Responses.J Immunol. 2021 Nov 15;207(10):2521-2533. doi: 10.4049/jimmunol.2100144. Epub 2021 Oct 13. J Immunol. 2021. PMID: 34645689 Free PMC article.
References
-
- Sanyal J, Chakraborty DP, Sarkar B, Banerjee TK, Mukherjee SC, Ray BC, Rao VR (2010) Environmental and familial risk factors of Parkinsons disease: case-control study. Can J Neurol Sci Le journal canadien des sciences neurologiques 37(5):637–642 - PubMed
-
- Pinto F, Mazza S (1971) Psychic symptoms in levodopa treatment of Parkinsons’ disease. II. Riv Neurobiol 17(2):155–159 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

