VCF-Server: A web-based visualization tool for high-throughput variant data mining and management
- PMID: 31127704
- PMCID: PMC6625089
- DOI: 10.1002/mgg3.641
VCF-Server: A web-based visualization tool for high-throughput variant data mining and management
Abstract
Background: Next-generation sequencing (NGS) has been widely used in both clinics and research. It has become the most powerful tool for diagnosing genetic disorders and investigating disease etiology through the discovery of genetic variants. Variants identified by NGS are stored in variant call format (VCF) files. However, querying and filtering VCF files are extremely difficult for researchers without programming skills. Furthermore, as the mutation data are increasing exponentially, there is an urgent need to develop tools to manage these variant data in a centralized way.
Methods: The VCF-Server was developed as a web-based visualization tool to support the interactive analysis of genetic variant data. It allows researchers and medical geneticists to manage, annotate, filter, query, and export variants in a fast and effective way.
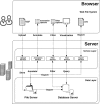
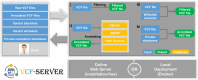
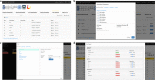
Results: In this study, we developed the VCF-Server, a powerful and easily accessible tool for researchers and medical geneticists to perform variant analysis. Users can query VCFs, annotate, and filter variants without knowing programming code. Once the VCF file is uploaded, VCF-Server allows users to annotate the VCF with commonly used databases or user-defined variant annotations (including variant blacklist and whitelist). Variant information in the VCF is shown visually via the interactive graphical interface. Users can filter the variants with flexible filtering rules, and the prioritized variants can be exported locally for further analysis. As VCF-Server adopts a web file system, files in the VCF-Server can be stored and managed in a centralized way. Moreover, VCF-Server allows direct web-based analysis (accessible through either desktop computers or mobile devices) as well as local deployment.
Conclusions: With an easy-to-use graphical interface, VCF-Server allows researchers with little bioinformatics background to explore and mine mutation data, which may broaden the application of NGS technology in clinics and research. The tool is freely available for use at https://www.diseasegps.org/VCF-Server?lan = eng.
Keywords: NGS; VCF visualization; software; variant filtering; variant management.
© 2019 The Authors. Molecular Genetics & Genomic Medicine published by Wiley Periodicals, Inc.
Conflict of interest statement
The authors declare that they have no competing interests. The VCF‐Server is freely available, but the copyrights of all the annotation databases on VCF‐Server belong to the original copyright owners or organizations. If used for commercial purpose, the user must acquire the license(s) of annotation database(s) for commercial entities.
Figures
Similar articles
-
VCF-Miner: GUI-based application for mining variants and annotations stored in VCF files.Brief Bioinform. 2016 Mar;17(2):346-51. doi: 10.1093/bib/bbv051. Epub 2015 Jul 25. Brief Bioinform. 2016. PMID: 26210358 Free PMC article.
-
VCF.Filter: interactive prioritization of disease-linked genetic variants from sequencing data.Nucleic Acids Res. 2017 Jul 3;45(W1):W567-W572. doi: 10.1093/nar/gkx425. Nucleic Acids Res. 2017. PMID: 28520890 Free PMC article.
-
DaMold: A data-mining platform for variant annotation and visualization in molecular diagnostics research.Hum Mutat. 2017 Jul;38(7):778-787. doi: 10.1002/humu.23227. Epub 2017 May 30. Hum Mutat. 2017. PMID: 28397319
-
vcfView: An Extensible Data Visualization and Quality Assurance Platform for Integrated Somatic Variant Analysis.Cancer Inform. 2020 Nov 11;19:1176935120972377. doi: 10.1177/1176935120972377. eCollection 2020. Cancer Inform. 2020. PMID: 33239857 Free PMC article. Review.
-
Open-Source Browser-Based Tools for Structure-Based Computer-Aided Drug Discovery.Molecules. 2022 Jul 20;27(14):4623. doi: 10.3390/molecules27144623. Molecules. 2022. PMID: 35889494 Free PMC article. Review.
Cited by
-
diseaseGPS: auxiliary diagnostic system for genetic disorders based on genotype and phenotype.Bioinformatics. 2023 Sep 2;39(9):btad517. doi: 10.1093/bioinformatics/btad517. Bioinformatics. 2023. PMID: 37647638 Free PMC article.
-
GenoSee: a novel visualization tool for graphical genotypes.Breed Sci. 2024 Dec;74(5):454-461. doi: 10.1270/jsbbs.24041. Epub 2024 Nov 23. Breed Sci. 2024. PMID: 39897665 Free PMC article.
-
Variant graph craft (VGC): a comprehensive tool for analyzing genetic variation and identifying disease-causing variants.BMC Bioinformatics. 2024 Sep 3;25(1):288. doi: 10.1186/s12859-024-05875-7. BMC Bioinformatics. 2024. PMID: 39227781 Free PMC article.
-
VIVA (VIsualization of VAriants): A VCF File Visualization Tool.Sci Rep. 2019 Sep 2;9(1):12648. doi: 10.1038/s41598-019-49114-z. Sci Rep. 2019. PMID: 31477778 Free PMC article.
-
VCFshiny: an R/Shiny application for interactively analyzing and visualizing genetic variants.Bioinform Adv. 2023 Aug 26;3(1):vbad107. doi: 10.1093/bioadv/vbad107. eCollection 2023. Bioinform Adv. 2023. PMID: 37701675 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

