Detection of Epidermal Growth Factor Receptor (EGFR) Gene Mutation in Formalin Fixed Paraffin Embedded Tissue by Polymerase Chain Reaction-Single Strand Conformational Polymorphism (PCR-SSCP) in Non-Small Cell Lung Cancer in the Northeastern Region of Thailand
- PMID: 31127887
- PMCID: PMC6857892
- DOI: 10.31557/APJCP.2019.20.5.1339
Detection of Epidermal Growth Factor Receptor (EGFR) Gene Mutation in Formalin Fixed Paraffin Embedded Tissue by Polymerase Chain Reaction-Single Strand Conformational Polymorphism (PCR-SSCP) in Non-Small Cell Lung Cancer in the Northeastern Region of Thailand
Abstract
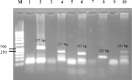
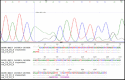
Background: The use of targeted specific genes in therapeutic and treatment decisions has been considered for lung cancer. The epidermal growth factor receptor (EGFR) gene, which is over expressed in non-small cell lung cancer (NSCLC), was considered as one of the targeted specific genes. EGFR mutations in exons 18–21, which encode a portion of the EGFR kinase domain, were found in NSCLC patients and were associated with the response of EGFRtyrosine kinase inhibitors (EGFR-TKIs). Therefore, a molecular technique for EGFR mutation detection has important benefits for therapy in NSCLC patients. This study aims to determine the EGFR mutations in patients with NSCLC using polymerase chain reaction-single strand conformational polymorphism (PCR-SSCP) in exons 18-21. Methods: DNA samples were extracted from formalin fixed paraffin embedded tissues of NSCLC patients who attended hospital. The extracted DNA was used as a template for the EGFR gene amplification. Results: Occurrence of EGFR mutations were found in 29 out of 50 cases (58%).The frequency of EGFR mutations by first PCR at exon 18, 19, 20 and 21 were 6 (12%), 19 (38%) 20 (40%) and at 21 (42%), respectively. By PCR-SSCP, the frequencies of EGFR mutations at exon 18, 19, 20 and 21 were 3(6%), 18(36%), 23(46%) and 13(26%), respectively. All of the mutations found were in agreement with DNA sequencings. Conclusion: The high frequency of EGFR mutations in NSCLC suggests that PCR-SSCP is a efficient screening method and useful for treatment plan.
Keywords: Epidermal growth factor receptor; polymerase chain reaction; single strand conformational polymorphism.
Creative Commons Attribution License
Figures






Similar articles
-
A universal method for the mutational analysis of K-ras and p53 gene in non-small-cell lung cancer using formalin-fixed paraffin-embedded tissue.Diagn Mol Pathol. 1995 Dec;4(4):266-73. doi: 10.1097/00019606-199512000-00007. Diagn Mol Pathol. 1995. PMID: 8634783
-
Screening for EGFR and KRAS mutations in non-small cell lung carcinomas using DNA extraction by hydrothermal pressure coupled with PCR-based direct sequencing.Int J Clin Exp Pathol. 2013 Aug 15;6(9):1880-9. eCollection 2013. Int J Clin Exp Pathol. 2013. PMID: 24040454 Free PMC article.
-
[Relationship of epidermal growth factor receptor gene mutations, clinicopathologic features and prognosis in patients with non-small cell lung cancer].Zhonghua Bing Li Xue Za Zhi. 2011 Oct;40(10):679-82. Zhonghua Bing Li Xue Za Zhi. 2011. PMID: 22321547 Chinese.
-
Is the epidermal growth factor receptor status in lung cancers reflected in clinicopathologic features?Arch Pathol Lab Med. 2010 Jan;134(1):66-72. doi: 10.5858/2008-0586-RAR1.1. Arch Pathol Lab Med. 2010. PMID: 20073607 Review.
-
[Studies and Progress of EGFR exon 20 Insertion Mutation in Non-small Cell Lung Cancer].Zhongguo Fei Ai Za Zhi. 2020 Feb 20;23(2):118-126. doi: 10.3779/j.issn.1009-3419.2020.02.07. Zhongguo Fei Ai Za Zhi. 2020. PMID: 32093456 Free PMC article. Review. Chinese.
Cited by
-
Application of the conventional and novel methods in testing EGFR variants for NSCLC patients in the last 10 years through different regions: a systematic review.Mol Biol Rep. 2021 Apr;48(4):3593-3604. doi: 10.1007/s11033-021-06379-w. Epub 2021 May 10. Mol Biol Rep. 2021. PMID: 33973139
-
A DNA based visual and colorimetric aggregation assay for the early growth factor receptor (EGFR) mutation by using unmodified gold nanoparticles.Mikrochim Acta. 2019 Jul 18;186(8):546. doi: 10.1007/s00604-019-3696-y. Mikrochim Acta. 2019. PMID: 31321546
-
Molecular alterations and clinical prognostic factors in resectable non-small cell lung cancer.BMC Cancer. 2024 Feb 13;24(1):200. doi: 10.1186/s12885-024-11934-2. BMC Cancer. 2024. PMID: 38347487 Free PMC article.
-
Expert recommendations for biomarker evaluation of advanced non-small cell lung cancer in Thailand.Transl Lung Cancer Res. 2025 Jul 31;14(7):2387-2402. doi: 10.21037/tlcr-2025-201. Epub 2025 Jul 28. Transl Lung Cancer Res. 2025. PMID: 40799428 Free PMC article. Review.
References
-
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide:sources, methods and major patterns in GLOBOCAN. Int J Cancer 2012. 2012;136:359–86. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

