Recreation of in-host acquired single nucleotide polymorphisms by CRISPR-Cas9 reveals an uncharacterised gene playing a role in Aspergillus fumigatus azole resistance via a non-cyp51A mediated resistance mechanism
- PMID: 31128273
- PMCID: PMC6876285
- DOI: 10.1016/j.fgb.2019.05.005
Recreation of in-host acquired single nucleotide polymorphisms by CRISPR-Cas9 reveals an uncharacterised gene playing a role in Aspergillus fumigatus azole resistance via a non-cyp51A mediated resistance mechanism
Abstract
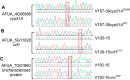
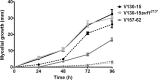
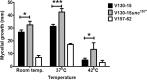
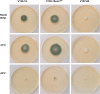
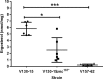
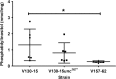
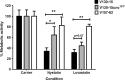
The human host comprises a range of specific niche environments. In order to successfully persist, pathogens such as Aspergillus fumigatus must adapt to these environments. One key example of in-host adaptation is the development of resistance to azole antifungals. Azole resistance in A. fumigatus is increasingly reported worldwide and the most commonly reported mechanisms are cyp51A mediated. Using a unique series of A. fumigatus isolates, obtained from a patient suffering from persistent and recurrent invasive aspergillosis over 2 years, this study aimed to gain insight into the genetic basis of in-host adaptation. Single nucleotide polymorphisms (SNPs) unique to a single isolate in this series, which had developed multi-azole resistance in-host, were identified. Two nonsense SNPs were recreated using CRISPR-Cas9; these were 213* in svf1 and 167* in uncharacterised gene AFUA_7G01960. Phenotypic analyses including antifungal susceptibility testing, mycelial growth rate assessment, lipidomics analysis and statin susceptibility testing were performed to associate genotypes to phenotypes. This revealed a role for svf1 in A. fumigatus oxidative stress sensitivity. In contrast, recapitulation of 167* in AFUA_7G01960 resulted in increased itraconazole resistance. Comprehensive lipidomics analysis revealed decreased ergosterol levels in strains containing this SNP, providing insight to the observed itraconazole resistance. Decreases in ergosterol levels were reflected in increased resistance to lovastatin and nystatin. Importantly, this study has identified a SNP in an uncharacterised gene playing a role in azole resistance via a non-cyp51A mediated resistance mechanism. This mechanism is of clinical importance, as this SNP was identified in a clinical isolate, which acquired azole resistance in-host.
Keywords: Aspergillus fumigatus; Azole resistance; CRISPR-Cas9; Ergosterol; In-host adaptation.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Figures

Similar articles
-
In-host microevolution of Aspergillus fumigatus: A phenotypic and genotypic analysis.Fungal Genet Biol. 2018 Apr;113:1-13. doi: 10.1016/j.fgb.2018.02.003. Epub 2018 Feb 23. Fungal Genet Biol. 2018. PMID: 29477713 Free PMC article.
-
High detection rate of azole-resistant Aspergillus fumigatus after treatment with azole antifungal drugs among patients with chronic pulmonary aspergillosis in a single hospital setting with low azole resistance.Med Mycol. 2021 Apr 6;59(4):327-334. doi: 10.1093/mmy/myaa052. Med Mycol. 2021. PMID: 32642756
-
Non-cyp51A Azole-Resistant Aspergillus fumigatus Isolates with Mutation in HMG-CoA Reductase.Emerg Infect Dis. 2018 Oct;24(10):1889-1897. doi: 10.3201/eid2410.180730. Emerg Infect Dis. 2018. PMID: 30226177 Free PMC article.
-
Azole resistance in Aspergillus fumigatus- comprehensive review.Arch Microbiol. 2024 Jun 15;206(7):305. doi: 10.1007/s00203-024-04026-z. Arch Microbiol. 2024. PMID: 38878211 Review.
-
Azole resistant Aspergillus fumigatus: an emerging problem.Med Mal Infect. 2013 Apr;43(4):139-45. doi: 10.1016/j.medmal.2013.02.010. Epub 2013 Apr 4. Med Mal Infect. 2013. PMID: 23562488 Review.
Cited by
-
Antifungal Activity of Antimicrobial Peptides and Proteins against Aspergillus fumigatus.J Fungi (Basel). 2020 May 18;6(2):65. doi: 10.3390/jof6020065. J Fungi (Basel). 2020. PMID: 32443413 Free PMC article.
-
Genomic Epidemiology Identifies Azole Resistance Due to TR34/L98H in European Aspergillus fumigatus Causing COVID-19-Associated Pulmonary Aspergillosis.J Fungi (Basel). 2023 Nov 13;9(11):1104. doi: 10.3390/jof9111104. J Fungi (Basel). 2023. PMID: 37998909 Free PMC article.
-
CRISPR/Cas9 gene editing: a novel strategy for fighting drug resistance in respiratory disorders.Cell Commun Signal. 2024 Jun 14;22(1):329. doi: 10.1186/s12964-024-01713-8. Cell Commun Signal. 2024. PMID: 38877530 Free PMC article. Review.
-
Genome-Wide Association and Selective Sweep Studies Reveal the Complex Genetic Architecture of DMI Fungicide Resistance in Cercospora beticola.Genome Biol Evol. 2021 Sep 1;13(9):evab209. doi: 10.1093/gbe/evab209. Genome Biol Evol. 2021. PMID: 34499119 Free PMC article.
-
Antifungal Exposure and Resistance Development: Defining Minimal Selective Antifungal Concentrations and Testing Methodologies.Front Fungal Biol. 2022 Jun 13;3:918717. doi: 10.3389/ffunb.2022.918717. eCollection 2022. Front Fungal Biol. 2022. PMID: 37746188 Free PMC article.
References
-
- Albarrag A.M., Anderson M.J., Howard S.J., Robson G.D., Warn P.A., Sanglard D., Denning D.W. Interrogation of related clinical pan-azole-resistant Aspergillus fumigatus strains: G138C, Y431C, and G434C single nucleotide polymorphisms in cyp51A, upregulation of cyp51A, and integration and activation of transposon Atf1 in the cyp51A promoter. Antimicrob. Agents Chemother. 2011;55:5113–5121. - PMC - PubMed
-
- Aleksenko A., Clutterbuck A.J. Autonomous Plasmid Replication in Aspergillus nidulans: AMA1 and MATE Elements. Fungal Genet. Biol. 1997;21:373–387. - PubMed
-
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

