MicroRNA-1305 Inhibits the Stemness of LCSCs and Tumorigenesis by Repressing the UBE2T-Dependent Akt-Signaling Pathway
- PMID: 31128423
- PMCID: PMC6535505
- DOI: 10.1016/j.omtn.2019.04.013
MicroRNA-1305 Inhibits the Stemness of LCSCs and Tumorigenesis by Repressing the UBE2T-Dependent Akt-Signaling Pathway
Abstract
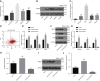
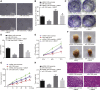
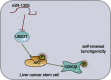
MicroRNAs (miRNAs) are involved in the maintenance of the cancer stem cell (CSC) phenotype by binding to genes and proteins that modulate cell proliferation and/or cell apoptosis. In our study, we aimed to investigate the role of miR-1305 in the proliferation and self-renewal of liver CSCs (LCSCs) via the ubiquitin-conjugating enzyme E2T (UBE2T)-mediated Akt-signaling pathway. Differentially expressed genes in human hepatocellular carcinoma (HCC) were obtained by in silico analysis. The relationship between miR-1305 and UBE2T was verified by dual luciferase reporter gene assay. qRT-PCR and western blot analysis were performed to determine the expression of UBE2T, the Akt-signaling pathway, and stemness-related factors in LCSCs. In addition, miR-1305 disrupted the activation of the Akt-signaling pathway by targeting UBE2T, and, ultimately, it repressed the sphere formation, colony formation, and proliferation, as well as tumorigenicity of LCSCs. In summary, miR-1305 targeted UBE2T to inhibit the Akt-signaling pathway, thereby suppressing the self-renewal and tumorigenicity of LCSCs. Those findings may provide an enhanced understanding of miR-1305 as a therapeutic target to limit the progression of LCSCs.
Keywords: Akt-signaling pathway; hepatocellular carcinoma; liver cancer stem cells; microRNA-1305; self-renewal; stemness; tumorigenicity; ubiquitin-conjugating enzyme E2T.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Figures




References
-
- Tang H., Jin Y., Jin S., Tan Z., Peng Z., Kuang Y. Arsenite inhibits the function of CD133+ CD13+ liver cancer stem cells by reducing PML and Oct4 protein expression. Tumour Biol. 2016;37:14103–14115. - PubMed
- Tang, H., Jin, Y., Jin, S., Tan, Z., Peng, Z., and Kuang, Y. (2016). Arsenite inhibits the function of CD133+ CD13+ liver cancer stem cells by reducing PML and Oct4 protein expression. Tumour Biol. 37, 14103-14115. - PubMed
-
- Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., Jemal A., Yu X.Q., He J. Cancer statistics in China, 2015. CA Cancer J. Clin. 2016;66:115–132. - PubMed
- Chen, W., Zheng, R., Baade, P.D., Zhang, S., Zeng, H., Bray, F., Jemal, A., Yu, X.Q., and He, J. (2016). Cancer statistics in China, 2015. CA Cancer J. Clin. 66, 115-132. - PubMed
-
- Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–E386. - PubMed
- Ferlay, J., Soerjomataram, I., Dikshit, R., Eser, S., Mathers, C., Rebelo, M., Parkin, D.M., Forman, D., and Bray, F. (2015). Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136, E359-E386. - PubMed
-
- Fong Z.V., Tanabe K.K. The clinical management of hepatocellular carcinoma in the United States, Europe, and Asia: a comprehensive and evidence-based comparison and review. Cancer. 2014;120:2824–2838. - PubMed
- Fong, Z.V., and Tanabe, K.K. (2014). The clinical management of hepatocellular carcinoma in the United States, Europe, and Asia: a comprehensive and evidence-based comparison and review. Cancer 120, 2824-2838. - PubMed
-
- Chen Y., Yu D., Zhang H., He H., Zhang C., Zhao W., Shao R.G. CD133(+)EpCAM(+) phenotype possesses more characteristics of tumor initiating cells in hepatocellular carcinoma Huh7 cells. Int. J. Biol. Sci. 2012;8:992–1004. - PMC - PubMed
- Chen, Y., Yu, D., Zhang, H., He, H., Zhang, C., Zhao, W., and Shao, R.G. (2012). CD133(+)EpCAM(+) phenotype possesses more characteristics of tumor initiating cells in hepatocellular carcinoma Huh7 cells. Int. J. Biol. Sci. 8, 992-1004. - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

