The small non-coding RNA profile of mouse oocytes is modified during aging
- PMID: 31128574
- PMCID: PMC6555462
- DOI: 10.18632/aging.101947
The small non-coding RNA profile of mouse oocytes is modified during aging
Abstract
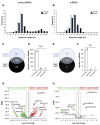
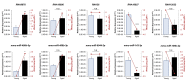
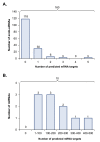
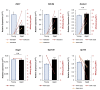
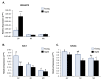
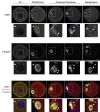
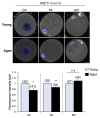
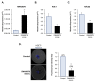
Oocytes are reliant on messenger RNA (mRNA) stores to support their survival and integrity during a protracted period of transcriptional dormancy as they await ovulation. Oocytes are, however, known to experience an age-associated alteration in mRNA transcript abundance, a phenomenon that contributes to reduced developmental potential. Here we have investigated whether the expression profile of small non-protein-coding RNAs (sRNAs) is similarly altered in aged mouse oocytes. The application of high throughput sequencing revealed substantial changes to the global sRNA profile of germinal vesicle stage oocytes from young (4-6 weeks) and aged mice (14-16 months). Among these, 160 endogenous small-interfering RNAs (endo-siRNAs) and 10 microRNAs (miRNAs) were determined to differentially accumulate within young and aged oocytes. Further, we revealed decreased expression of two members of the kinesin protein family, Kifc1 and Kifc5b, in aged oocytes; family members selectively targeted for expression regulation by endo-siRNAs of elevated abundance. The implications of reduced Kifc1 and Kifc5b expression were explored using complementary siRNA-mediated knockdown and pharmacological inhibition strategies, both of which led to increased rates of aneuploidy in otherwise healthy young oocytes. Collectively, our data raise the prospect that altered sRNA abundance, specifically endo-siRNA abundance, could influence the quality of the aged oocyte.
Keywords: aneuploidy; endo-siRNA; kinesin; maternal aging; meiosis; miRNA; oocyte; small non-coding RNA.
Conflict of interest statement
Figures
Similar articles
-
Essential Role for endogenous siRNAs during meiosis in mouse oocytes.PLoS Genet. 2015 Feb 19;11(2):e1005013. doi: 10.1371/journal.pgen.1005013. eCollection 2015 Feb. PLoS Genet. 2015. PMID: 25695507 Free PMC article.
-
The maternal-to-zygotic transition in bovine in vitro-fertilized embryos is associated with marked changes in small non-coding RNAs†.Biol Reprod. 2019 Feb 1;100(2):331-350. doi: 10.1093/biolre/ioy190. Biol Reprod. 2019. PMID: 30165428
-
Endo-siRNA deficiency results in oocyte maturation failure and apoptosis in porcine oocytes.Reprod Fertil Dev. 2017 Oct;29(11):2168-2174. doi: 10.1071/RD16498. Reprod Fertil Dev. 2017. PMID: 28399989
-
Centrosome and microtubule functions and dysfunctions in meiosis: implications for age-related infertility and developmental disorders.Reprod Fertil Dev. 2015 Jul;27(6):934-43. doi: 10.1071/RD14493. Reprod Fertil Dev. 2015. PMID: 25903261 Review.
-
Small RNAs derived from longer non-coding RNAs.Biochimie. 2011 Nov;93(11):1905-15. doi: 10.1016/j.biochi.2011.07.032. Epub 2011 Aug 9. Biochimie. 2011. PMID: 21843590 Review.
Cited by
-
Common factors among three types of cells aged in mice.Biogerontology. 2023 Jun;24(3):363-375. doi: 10.1007/s10522-023-10035-0. Epub 2023 Apr 21. Biogerontology. 2023. PMID: 37081236
-
The dynamics of DNA methylation, histone methylation and acetylation during oocyte aging in mammalian species and possible interventions to regulate them.J Assist Reprod Genet. 2025 Jul 14. doi: 10.1007/s10815-025-03577-4. Online ahead of print. J Assist Reprod Genet. 2025. PMID: 40653579 Review.
-
Ovarian aging: energy metabolism of oocytes.J Ovarian Res. 2024 May 31;17(1):118. doi: 10.1186/s13048-024-01427-y. J Ovarian Res. 2024. PMID: 38822408 Free PMC article. Review.
-
MicroRNA-Mediated Gene Regulatory Mechanisms in Mammalian Female Reproductive Health.Int J Mol Sci. 2021 Jan 19;22(2):938. doi: 10.3390/ijms22020938. Int J Mol Sci. 2021. PMID: 33477832 Free PMC article. Review.
-
Mouse Oocytes, A Complex Single Cell Transcriptome.Front Cell Dev Biol. 2022 Mar 7;10:827937. doi: 10.3389/fcell.2022.827937. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35321242 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

