Live-Animal Epigenome Editing: Convergence of Novel Techniques
- PMID: 31128888
- PMCID: PMC6608710
- DOI: 10.1016/j.tig.2019.04.007
Live-Animal Epigenome Editing: Convergence of Novel Techniques
Abstract
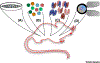
Epigenome editing refers to the generation of precise chromatin alterations and their effects on gene expression and cell biology. Until recently, much of the efforts in epigenome editing were limited to tissue culture models of disease. However, the convergence of techniques from different fields including mammalian genetics, virology, and CRISPR engineering is advancing epigenome editing into a new era. Researchers are increasingly embracing the use of multicellular model organisms to test the role of specific chromatin alterations in development and disease. The challenge of successful live-animal epigenomic editing will depend on a well-informed foundation of the current methodologies for cell-specific delivery and editing accuracy. Here we review the opportunities for basic research and therapeutic applications.
Keywords: CRISPR; animal models; cancer; delivery; epigenomic editing; neurodevelopment.
Copyright © 2019 Elsevier Ltd. All rights reserved.
Figures


References
-
- Wang L et al. (2018) Meganuclease targeting of PCSK9 in macaque liver leads to stable reduction in serum cholesterol. Nat. Biotechnol 36, 717–725 - PubMed
-
- Chiu A and Rao MS (2003) Human Embryonic Stem Cells, Springer Science & Business Media.

